
INTERGROWTH-21st Body Composition Equations
Source:vignettes/articles/ig-nbs-body-composition.Rmd
ig-nbs-body-composition.RmdThough Villar et al. (2017) published centiles for body
composition (fat mass, body fat percentage, and fat-free mass) in
newborns from the INTERGROWTH-21st project, they did not
publish the parameters of the models used to generate their centiles. To
implement these standards in gigs, we re-derived these
values, in this case using the published data to work back to linear
equations.
Contextual clues
To identify the way in which these body composition standards were
modelled, we need to look into how the published centile tables are
formatted. Consider the standard for fat mass for gestational age
("fmfga") in females:
fmfga_female#> # A tibble: 5 × 6
#> gest_days P03 P10 P50 P90 P97
#> <int> <int> <int> <int> <int> <int>
#> 1 266 29 123 324 525 619
#> 2 273 62 156 357 558 652
#> 3 280 96 190 391 592 686
#> 4 287 127 221 422 623 717
#> 5 294 150 244 445 646 740Differences between each GA in fmfga
Let’s first compare the differences over time in fat-mass for GA. In
each column, we’ll subtract the value of the previous row from the
current row to produce a table of differences (see
check_differences() below). Then we’ll check that the
differences in each row are equal for all centiles, and store this in
the all_equal column.
check_differences <- function(centile_tbl) {
centile_tbl |>
dplyr::mutate(dplyr::across(.cols = tidyselect::starts_with(match = "P"),
.fns = list(diff = \(x) x - dplyr::lag(x))),
.keep = "unused") |>
dplyr::filter(dplyr::if_any(
tidyselect::starts_with(match = "P"), ~ !is.na(.))) |>
dplyr::rowwise() |>
dplyr::mutate(
all_equal = dplyr::n_distinct(P03_diff, P10_diff, P50_diff, P90_diff,
P97_diff, na.rm = TRUE),
all_equal = as.logical(all_equal)) |>
dplyr::ungroup()
}Let’s check the results for fmfga_female:
check_differences(fmfga_female)#> # A tibble: 4 × 7
#> gest_days P03_diff P10_diff P50_diff P90_diff P97_diff all_equal
#> <int> <int> <int> <int> <int> <int> <lgl>
#> 1 273 33 33 33 33 33 TRUE
#> 2 280 34 34 34 34 34 TRUE
#> 3 287 31 31 31 31 31 TRUE
#> 4 294 23 23 23 23 23 TRUEThe differences in fat mass are equal across percentiles at all
gestational ages, according to the all_equal column. If the
standard deviation
()
changed with gestational age (GA), then all_equal would not
be at each age. This hints that for these centile curves, the mean value
()
for fat mass was calculated first, then summed with the product of the
desired
-score/centile
with a standard deviation
()
which is constant across the different GAs:
In the other standards
Does this equality hold for the other centile tables?
other_standards <- list(fmfga_male, bfpfga_male, bfpfga_female, ffmfga_male,
ffmfga_female)
vapply(X = other_standards,
FUN = \(x) all(check_differences(x)$all_equal),
FUN.VALUE = logical(length = 1L))#> [1] FALSE TRUE TRUE TRUE TRUENo. The first value, corresponding to
fmfga_male, is FALSE. Let’s see what the
differences look like for this table:
check_differences(fmfga_male)#> # A tibble: 4 × 7
#> gest_days P03_diff P10_diff P50_diff P90_diff P97_diff all_equal
#> <int> <int> <int> <int> <int> <int> <lgl>
#> 1 273 NA 38 39 39 39 FALSE
#> 2 280 40 40 40 40 40 TRUE
#> 3 287 42 42 42 42 42 TRUE
#> 4 294 24 24 24 24 24 TRUEThe issue here is a value of 38 rather than the
39s in the P10_diff column - but this
difference is small enough to be a consequence of rounding, as opposed
to a real difference in
for this GA compared to the other GAs.
Conclusion
To reconstruct these standards, we need to determine the equations for in each standard. We also need to calculate , which does not change with GA.
Extracting models from published tables
Custom functions
The following custom functions perform several tasks:
-
run_LMs()fits models to each INTERGROWTH-21st normative body composition standard usinglm(), then returns the fitted models in a list. -
std_deviation()determines for a given centile. -
extract_SDs()gets the standard deviations for each centile at each GA. -
extract_eqn()uses the above functions to extract a ‘best possible’ equation for the standard in use, then prints this to the console.
run_LMs <- function(df) {
model_linear <- lm(P50 ~ gest_days, data = df)
model_poly1 <- lm(P50 ~ poly(gest_days, degree = 1, raw = TRUE), data = df)
model_poly2 <- lm(P50 ~ poly(gest_days, degree = 2, raw = TRUE), data = df)
model_poly3 <- lm(P50 ~ poly(gest_days, degree = 3, raw = TRUE), data = df)
list(model_linear, model_poly1, model_poly2, model_poly3)
}
#' Extract standard deviation from centiles
#' @param P50 Value of y at mean
#' @param PX Value of y at centile specified by `p`
#' @param p Centile at which y = PX
std_deviation <- function(P50, PX, p) (PX - P50) / qnorm(p = p)
extract_SDs <- function(df) {
stddev_03 <- std_deviation(df$P50, df$P03, p = 0.03)
stddev_10 <- std_deviation(df$P50, df$P10, p = 0.10)
stddev_90 <- std_deviation(df$P50, df$P90, p = 0.90)
stddev_97 <- std_deviation(df$P50, df$P97, p = 0.97)
stddevs <- c(stddev_03, stddev_10, stddev_90, stddev_97)
mean(stddevs, na.rm = TRUE)
}
#' Extract equation and standard deviation; print to console
#' @param df A centile table for `fmfga`, `bfpfga` or `ffmfga`.
extract_eqn <- function(df, verbose = TRUE) {
li_models <- run_LMs(df)
r_sq_adj <- vapply(X = li_models,
FUN = \(x) summary(x)$adj.r.squared,
FUN.VALUE = numeric(length = 1L))
chosen_model <- li_models[[which(r_sq_adj == max(r_sq_adj))]]
if (!verbose) {
return(chosen_model)
}
model_coeffs <- coef(chosen_model)
coef_str <- lapply(
X = 2:length(model_coeffs),
FUN = \(x) {
start_str <- if (sign(model_coeffs[x]) == -1) "" else "+"
end_str <- if (x == 2) "* x" else paste0("* x^", x - 1)
paste(start_str, round(model_coeffs[x], 5), end_str)
}) |>
paste(collapse = " ")
sign_ <- if (sign(model_coeffs[2]) == -1) "" else "+"
cat(paste("y =", round(model_coeffs[[1]], 5), sign_, coef_str), sep = "\n")
SDs <- extract_SDs(df)
cat(paste("sigma =", round(mean(SDs, na.rm = TRUE), digits = 4)), sep = "\n")
invisible(chosen_model)
}Fat mass
Males
fmfga_male <- gigs::ig_nbs$fmfga$male$centiles
extract_eqn(fmfga_male)#> y = 96787.8 -1056.77381 * x + 3.83673 * x^2 -0.00462 * x^3
#> sigma = 152.0728Females
fmfga_female <- gigs::ig_nbs$fmfga$female$centiles
extract_eqn(fmfga_female)#> y = 44432.51429 -490.7602 * x + 1.80321 * x^2 -0.00219 * x^3
#> sigma = 156.8449Body fat percentage
Males
bfpfga_male <- gigs::ig_nbs$bfpfga$male$centiles
extract_eqn(bfpfga_male)#> y = 1521.84857 -16.62398 * x + 0.0605 * x^2 -7e-05 * x^3
#> sigma = 3.6562Females
bfpfga_female <- gigs::ig_nbs$bfpfga$female$centiles
extract_eqn(bfpfga_female)#> y = -66.09143 + + 0.48102 * x -0.00073 * x^2
#> sigma = 3.9547Fat-free mass
Males
ffmfga_male <- gigs::ig_nbs$ffmfga$male$centiles
extract_eqn(ffmfga_male)#> y = 347879.97143 -3780.52721 * x + 13.73032 * x^2 -0.01652 * x^3
#> sigma = 276.5094Females
ffmfga_female <- gigs::ig_nbs$ffmfga$female$centiles
extract_eqn(ffmfga_female)#> y = 154457.42857 -1689.0085 * x + 6.20845 * x^2 -0.00753 * x^3
#> sigma = 261.0625Comparing fitted values with published values
All plotting code for this section can be found in the source code for this vignette.
Reconstructing the equations in R
First, we will bind the coefficients from each model into a table. This table can then be used to look up the coefficients for each body composition standard.
all_standards <- list(fmfga_male, fmfga_female, bfpfga_male, bfpfga_female,
ffmfga_male, ffmfga_female)
models <- lapply(all_standards, FUN = \(standard) {
model_coeffs <- standard |>
extract_eqn(verbose = FALSE) |>
coef() |>
t() |>
as.data.frame()
if (ncol(model_coeffs) == 3) model_coeffs$extra <- 0
names(model_coeffs) <- c("intercept", "x", "x^2", "x^3")
model_coeffs$SD <- extract_SDs(standard)
model_coeffs
}) |>
do.call(what = "rbind")
rownames(models) <- c("fmfga_M" , "fmfga_F" , "bfpfga_M", "bfpfga_F",
"ffmfga_M", "ffmfga_F")
models#> intercept x x^2 x^3 SD
#> fmfga_M 96787.80000 -1056.7738095 3.836734694 -0.0046161322 152.072842
#> fmfga_F 44432.51429 -490.7602041 1.803206997 -0.0021865889 156.844908
#> bfpfga_M 1521.84857 -16.6239796 0.060495627 -0.0000728863 3.656170
#> bfpfga_F -66.09143 0.4810204 -0.000728863 0.0000000000 3.954716
#> ffmfga_M 347879.97143 -3780.5272109 13.730320700 -0.0165208941 276.509412
#> ffmfga_F 154457.42857 -1689.0085034 6.208454810 -0.0075315841 261.062458The body_comp_centile2value() will estimate
y for a given gestational age in days (x),
sex and acronym. The
body_comp_centile2value_old() function does the same, but
uses parameters from our older linear models for each standard.
ig_nbs_bc_centile2value <- function(p, x, sex, acronym) {
params <- t(models)[, paste0(acronym, "_", sex)]
SD <- params[[5]]
mu <- params[[1]] + params[[2]] * x + params[[3]] * x^2 + params[[4]] * x^3
mu + qnorm(p) * SD
}
ig_nbs_bc_centile2value_old <- function(p, x, sex, acronym) {
z <- qnorm(p)
if (sex == "M") {
switch(acronym,
"fmfga" = -1134.2 + 37.2 * x/7 + z * 152.1593,
"bfpfga" = -17.68 + 0.69 * x/7 + z * 3.6674,
"ffmfga" = -2487.6 + 139.9 * x/7 + z * 276.2276,
stop("Bad acronym", .call = F))
} else if (sex == "F") {
switch(acronym,
"fmfga" = -840.2 + 30.7 * x/7 + z * 156.8411,
"bfpfga" = -9.02 + 0.51 * x/7 + z * 3.9405,
"ffmfga" = -1279 + 105.3 * x/7 + z * 260.621,
stop("Bad acronym", .call = F))
}
}Comparing fitted values with published values
Making percentile tables
#' Replicate centile tables as presented by Villar et al.
repl_centile_tbls <- function(sex, acronym, bodycomp_fn) {
centiles <- c(0.03, 0.1, 0.5, 0.9, 0.97) |>
purrr::set_names(c("P03", "P10", "P50", "P90", "P97"))
tbl <- purrr::map(.x = centiles, .f = ~ {
p <- .x
column <- seq(266, 294, by = 7) |>
purrr::set_names(paste(1:5)) |>
purrr::map_dfc(.f = ~ {
bodycomp_fn(p, .x, sex, acronym)
}) |>
t()
column[which(column <= 0)] <- NA_real_
as.data.frame(column)
})
tbl <- suppressMessages(suppressWarnings(dplyr::bind_cols(tbl)))
colnames(tbl) <- names(centiles)
dplyr::mutate(tbl, gest_days = (38:42 * 7), .before = P03)
}
#' Make centile tables based on parameters from new polynomial models
repl_centile_tbls_new <- function(sex, acronym) {
repl_centile_tbls(sex, acronym, ig_nbs_bc_centile2value)
}
#' Make centile tables based on parameters from linear models
repl_centile_tbls_old <- function(sex, acronym) {
repl_centile_tbls(sex, acronym, ig_nbs_bc_centile2value_old)
}Observed vs fitted values
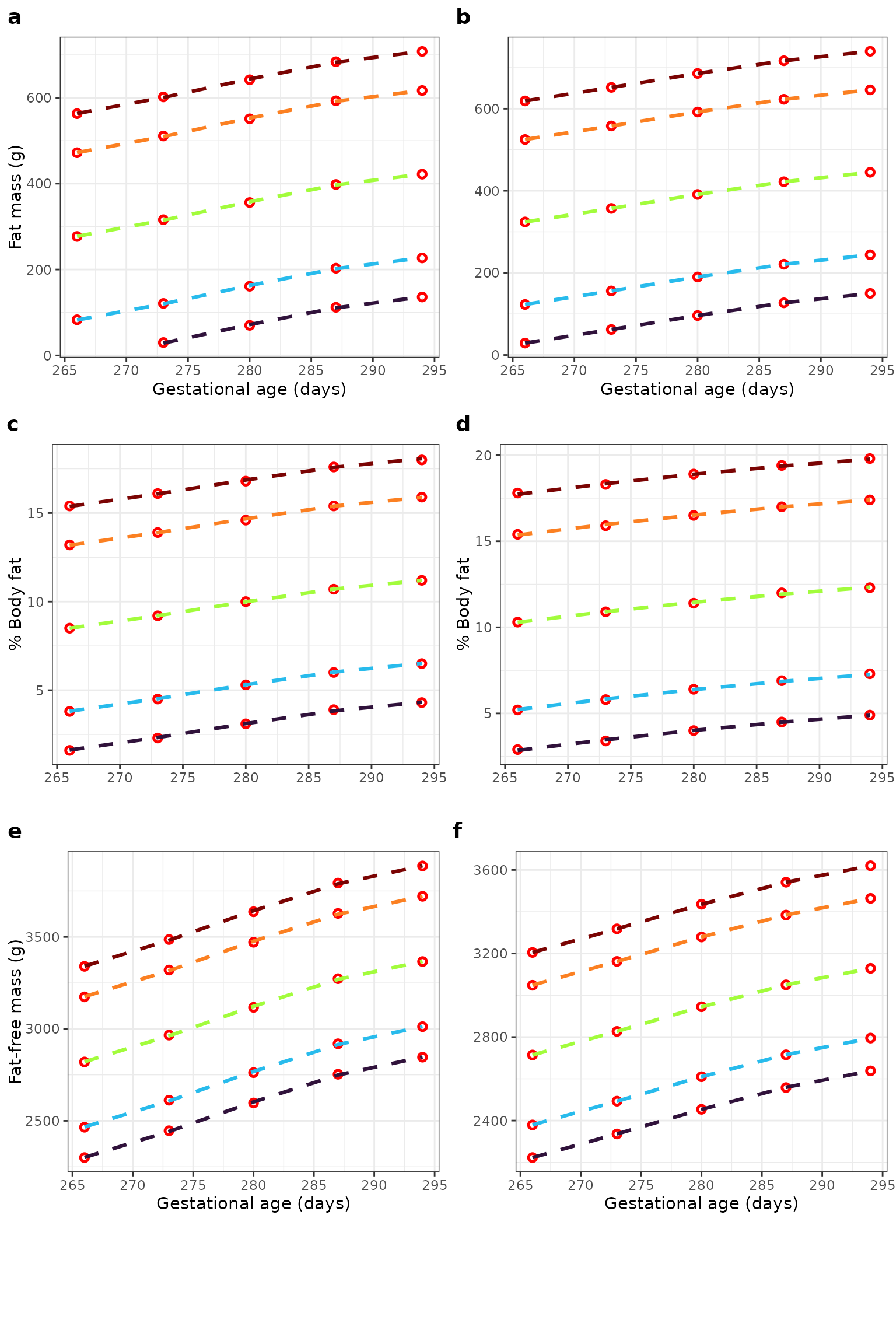
Observed and estimated normative centiles for body composition measures according to gestational age and sex. Centiles (3rd, 10th, 50th, 90th, and 97th) for fat mass (a males; b females); body fat percentage (c males; d females); and fat-free mass (e males; f females) according to gestational age. Red circles show the observed value for that centile and gestational age; dashed lines show estimates from linear equations fitted to these observed values.
Bland-Altman plots
Reference vs. predicted values
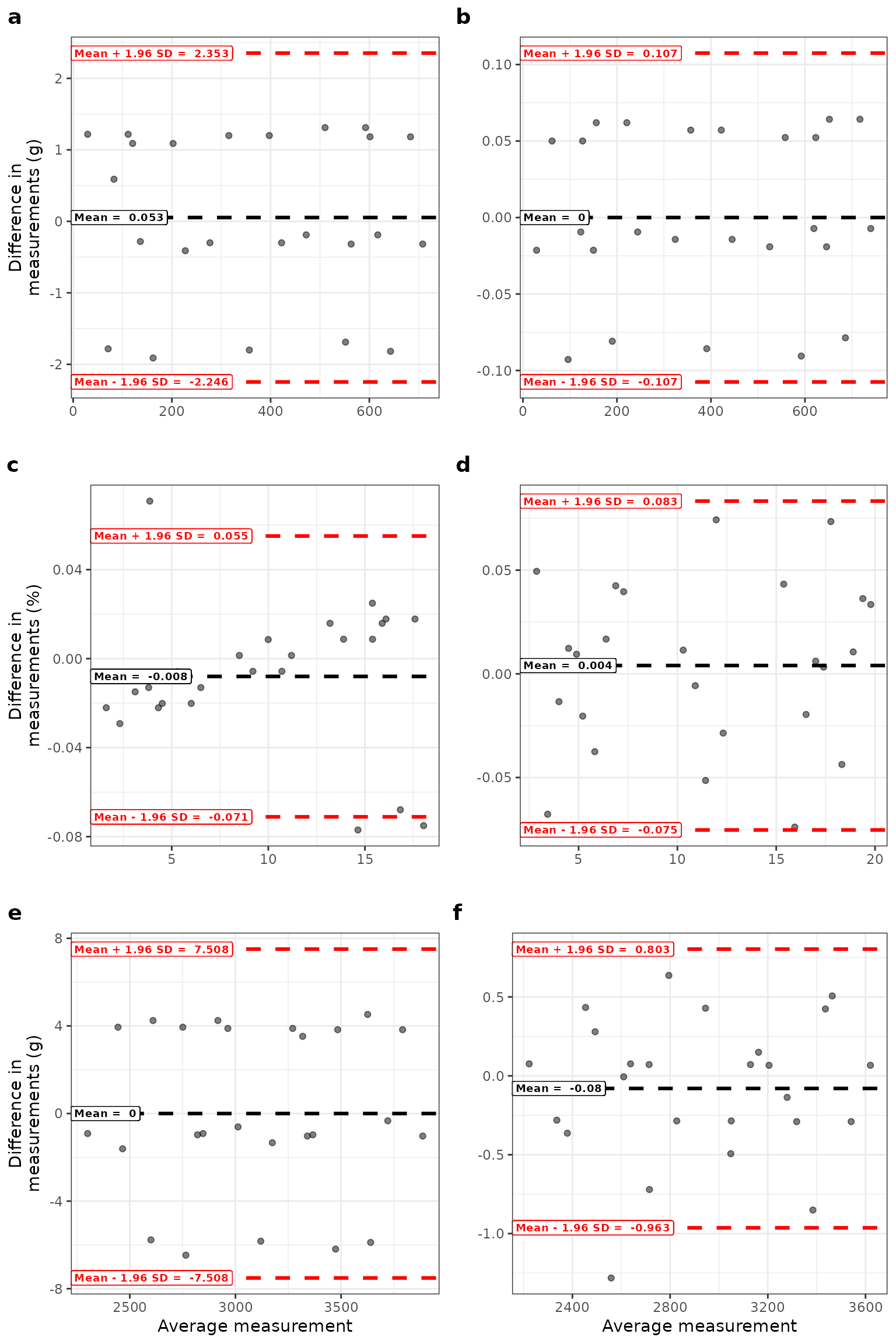
Bland-Altman plots for observed and estimated centile values for body composition measures according to gestational age and sex. Circles show differences between observed and estimated values for fat mass (a males; b females); body fat percentage (c males; d females); and fat-free mass (e males; f females) according to the average of the two measurements.
References
Villar J, Puglia FA, Fenton TR, Ismal LC, Staines-Urias E, Giuliani F, et al. Body composition at birth and its relationship with neonatal anthropometric ratios: the newborn body composition study of the INTERGROWTH-21st project Pediatric Research 2017, 82:305-316. doi: 10.1038/pr.2017.52