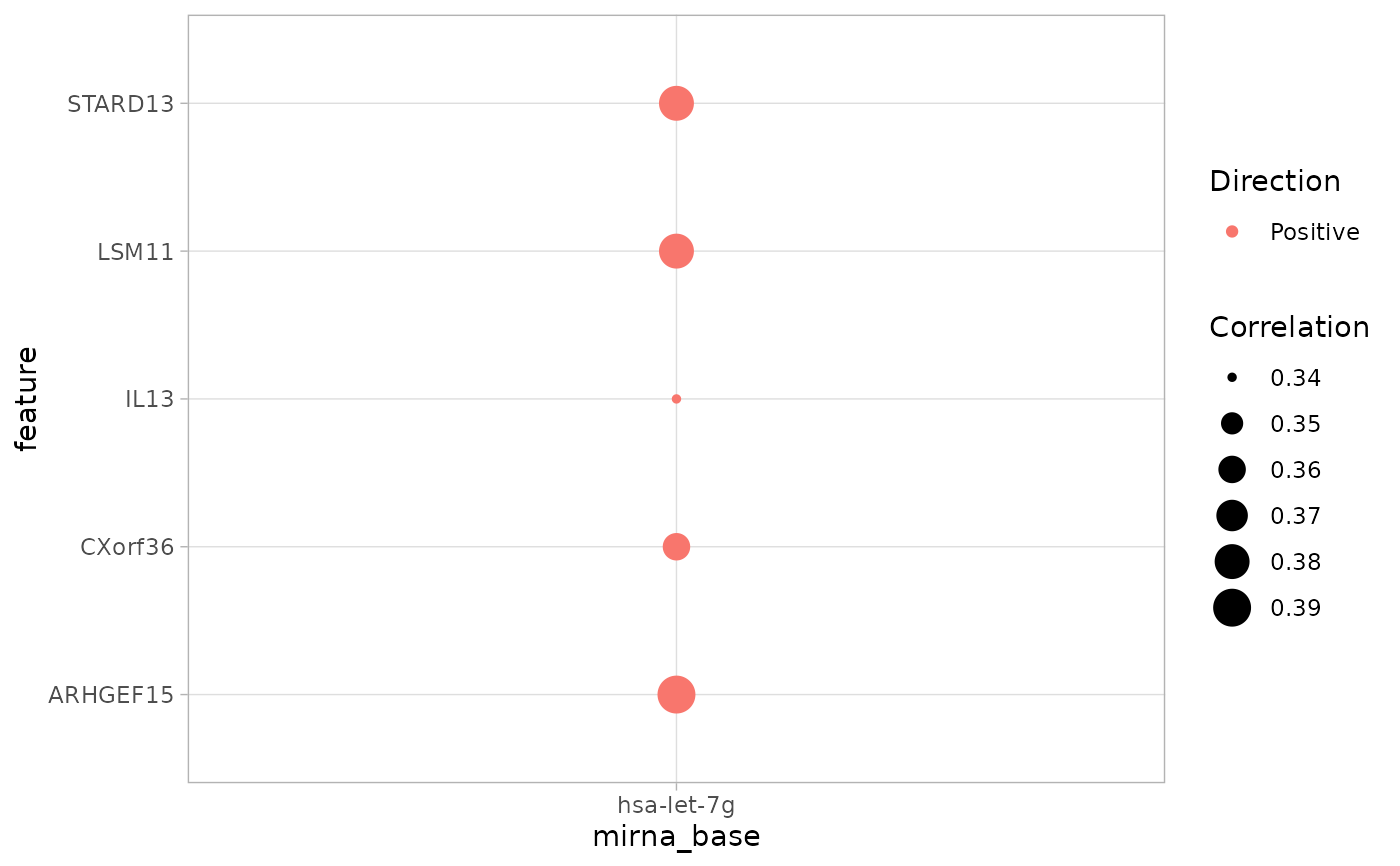
A dot plot of microRNA/TF correlation in a single study of TCGA. When the
object cmicroRNA/cTF contains more than one TCGA studies, the
argument study is a requirement.
Arguments
- ob
A cmicroRNA or cTF object such as this returned by calling cmicroRNA or cTF.
- study
A
charactervector of The Cancer Genome Atlas (TCGA) study identifiers. To view the available studies in TCGA project, https://tcga-data.nci.nih.gov/docs/publications/tcga. When left to defaultNULLall available studies will be included.- ...
Other options
Value
A ggplot object of a dot plot of the correlation values
between genes and microRNAs or transcription factors in a TCGA study.
Examples
# locate the testset file and connect
fl <- system.file('extdata', 'cRegulome.db', package = 'cRegulome')
conn <- RSQLite::dbConnect(RSQLite::SQLite(), fl)
# enter a custom query with different arguments
dat <- get_mir(conn,
mir = 'hsa-let-7g',
study = 'STES',
min_abs_cor = .3,
max_num = 5)
# make a cmicroRNA object
cmir <- cmicroRNA(dat)
# print object
cor_plot(cmir)