
Benchmarking Sparse Matrix Market Read Operations
Rohit Goswami
2023-11-03
Source:vignettes/fmm_read_bench.Rmd
fmm_read_bench.RmdIntroduction
This vignette demonstrates a benchmark comparing the
readMM function from the Matrix package
against the read_fmm function from the
fastMatMR package. Since Matrix does not
support reading or writing dense matrices, we focus on the sparse
case.
Benchmarking with Fixed Sparsity
We first benchmark for varying matrix sizes with fixed sparsity.
# Function to create a sparse matrix of given size
create_sparse_matrix <- function(n, sparsity = 0.7) {
mat <- matrix(0, nrow = n, ncol = n)
for (i in 1:n) {
for (j in 1:n) {
if (runif(1) > sparsity) {
mat[i, j] <- rnorm(1)
}
}
}
return(Matrix(mat, sparse = TRUE))
}
# Define a range of matrix sizes
sizes <- c(10, 100, 500, 1000, 2000, 3000)
# Prepare data frame to store results
results_fixed_sparsity <- data.frame()
# Benchmarking
for (n in sizes) {
message("Benchmarking for matrix size: ", n, "x", n)
# Generate a sparse matrix of size n x n
testmat <- create_sparse_matrix(n)
write_fmm(testmat, "sparse.mtx")
# Run the benchmarks, we coerce to a sparse matrix for readMM for fairness
bm <- microbenchmark(
Matrix_readMM = as(readMM("sparse.mtx"), "CsparseMatrix"),
fastMatMR_read_fmm = fmm_to_sparse_Matrix("sparse.mtx"),
times = 10
)
bm$size <- n
results_fixed_sparsity <- rbind(results_fixed_sparsity, bm)
}
#> Benchmarking for matrix size: 10x10
#> Benchmarking for matrix size: 100x100
#> Benchmarking for matrix size: 500x500
#> Benchmarking for matrix size: 1000x1000
#> Benchmarking for matrix size: 2000x2000
#> Benchmarking for matrix size: 3000x3000This is shown visually represented below:
# Plotting
suppressWarnings(print(
ggplot(results_fixed_sparsity, aes(x = size, y = time, color = expr)) +
geom_point() +
geom_smooth(method = "loess") +
ggtitle("Benchmarking reads with fixed sparsity for 70% sparsity") +
xlab("Matrix Size") +
ylab("Time (ns)")
))
#> `geom_smooth()` using formula = 'y ~ x'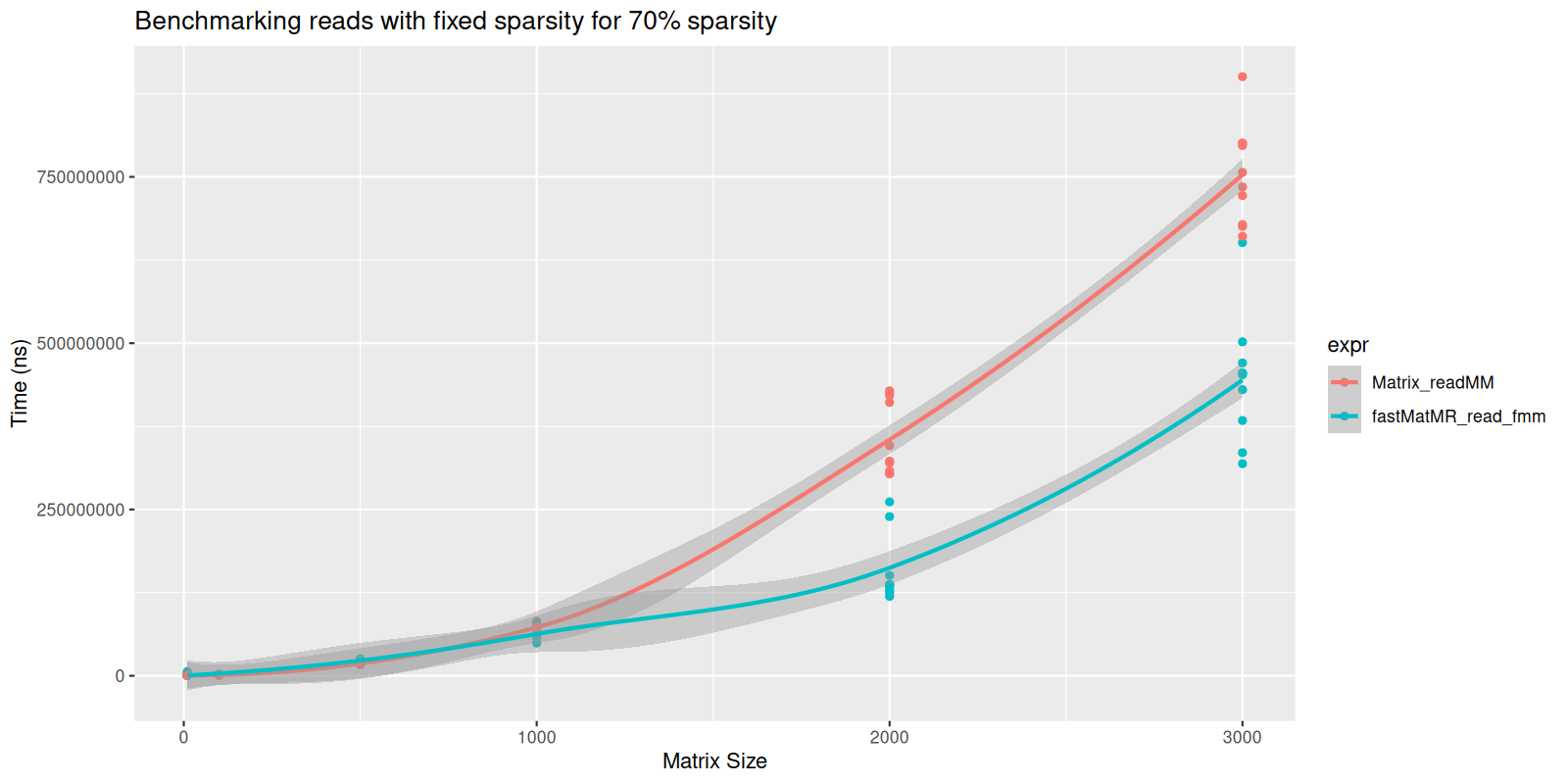
plot of chunk fixed-sparse-read
Benchmarking with Varying Sparsity
Now, we benchmark for varying sparsity patterns on a large matrix.
# Sparsity levels to test
sparsity_levels <- seq(0.45, 0.95, by = 0.1)
# Prepare data frame to store results
results_varying_sparsity <- data.frame()
# Benchmarking
for (sparsity in sparsity_levels) {
message("Benchmarking for sparsity level: ", sparsity)
# Generate a sparse matrix of size 2000 x 2000 with varying sparsity
testmat <- create_sparse_matrix(2000, sparsity)
write_fmm(testmat, "sparse.mtx")
# Run the benchmarks
bm <- microbenchmark(
Matrix_readMM = as(readMM("sparse.mtx"), "CsparseMatrix"),
fastMatMR_read_fmm = fmm_to_sparse_Matrix("sparse.mtx"),
times = 10
)
bm$sparsity <- sparsity
results_varying_sparsity <- rbind(results_varying_sparsity, bm)
}
#> Benchmarking for sparsity level: 0.45
#> Benchmarking for sparsity level: 0.55
#> Benchmarking for sparsity level: 0.65
#> Benchmarking for sparsity level: 0.75
#> Benchmarking for sparsity level: 0.85
#> Benchmarking for sparsity level: 0.95Now we can plot this:
ggplot(results_varying_sparsity, aes(x = sparsity, y = time, color = expr)) +
geom_point() +
geom_smooth(method = "loess") +
scale_x_log10() +
scale_y_log10() +
ggtitle("Benchmarking reads with varying sparsity for 2000 entries") +
xlab("Sparsity Level (log10)") +
ylab("Time (ns, log10)")
#> `geom_smooth()` using formula = 'y ~ x'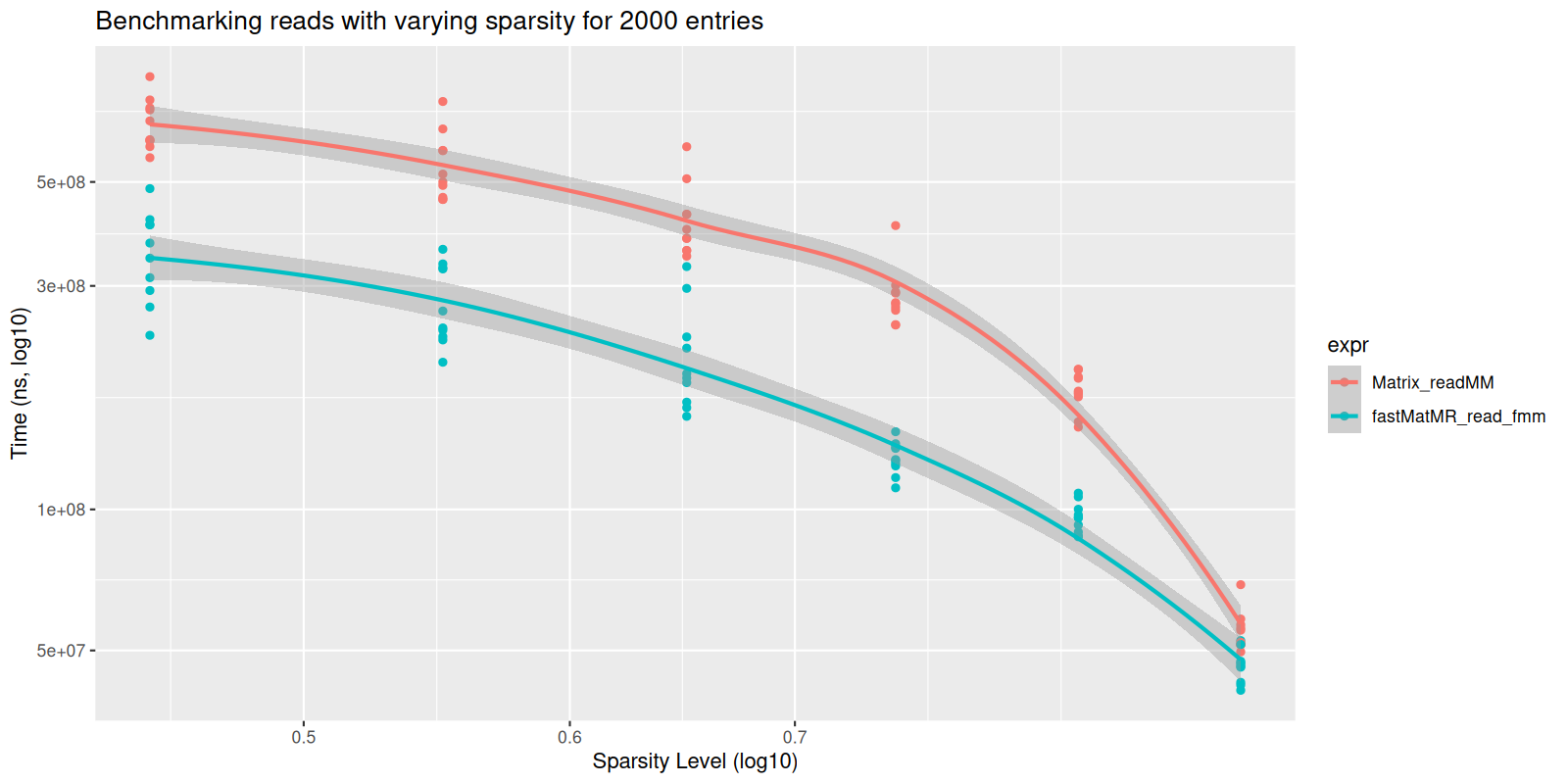
plot of chunk varying-sparse-read
Conclusions
We see that though there are no statistically significant differences
in speed for small matrices, the fastMatMR package is
significantly faster for large matrices. This is because the
readMM function from the Matrix reads data
into a triplet form, which gets slower for larger matrices.
Session Info
This vignette was computed in advance, with the corresponding session info:
sessionInfo()
#> R version 4.3.1 (2023-06-16)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Arch Linux
#>
#> Matrix products: default
#> BLAS: /usr/lib/libblas.so.3.11.0
#> LAPACK: /usr/lib/liblapack.so.3.11.0
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Iceland
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggplot2_3.4.4 microbenchmark_1.4.10 Matrix_1.5-4.1
#> [4] fastMatMR_1.2.5 testthat_3.1.10
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.4 xfun_0.40 htmlwidgets_1.6.2 devtools_2.4.5
#> [5] remotes_2.4.2.1 processx_3.8.2 lattice_0.21-8 callr_3.7.3
#> [9] generics_0.1.3 vctrs_0.6.3 tools_4.3.1 ps_1.7.5
#> [13] parallel_4.3.1 tibble_3.2.1 fansi_1.0.4 highr_0.10
#> [17] pkgconfig_2.0.3 desc_1.4.2 lifecycle_1.0.3 farver_2.1.1
#> [21] compiler_4.3.1 stringr_1.5.0 brio_1.1.3 munsell_0.5.0
#> [25] decor_1.0.2 httpuv_1.6.11 htmltools_0.5.6 usethis_2.2.2
#> [29] later_1.3.1 pillar_1.9.0 crayon_1.5.2 urlchecker_1.0.1
#> [33] ellipsis_0.3.2 cachem_1.0.8 sessioninfo_1.2.2 nlme_3.1-162
#> [37] mime_0.12 commonmark_1.9.0 tidyselect_1.2.0 digest_0.6.33
#> [41] stringi_1.7.12 dplyr_1.1.2 purrr_1.0.2 labeling_0.4.3
#> [45] splines_4.3.1 rprojroot_2.0.3 fastmap_1.1.1 grid_4.3.1
#> [49] colorspace_2.1-0 cli_3.6.1 magrittr_2.0.3 pkgbuild_1.4.2
#> [53] utf8_1.2.3 withr_2.5.0 prettyunits_1.1.1 scales_1.2.1
#> [57] promises_1.2.1 cpp11_0.4.6 roxygen2_7.2.3 memoise_2.0.1
#> [61] shiny_1.7.5 evaluate_0.21 knitr_1.43 miniUI_0.1.1.1
#> [65] mgcv_1.8-42 profvis_0.3.8 rlang_1.1.1 Rcpp_1.0.11
#> [69] xtable_1.8-4 glue_1.6.2 xml2_1.3.5 pkgload_1.3.2.1
#> [73] rstudioapi_0.15.0 R6_2.5.1 fs_1.6.3