Processing
Processing encapsulates the operations applied to satellite images to solve particular issues derived from errors, anomalies or discrepancies from sensors. For instance, cloud removal or sensor failures can lead to data gaps in time-series of satellite images. Additionally, noise from aerosols, dust, and sensor measurement errors can reduce the quality of the remotely sensed data. The Interpolation of Mean Anomalies (IMA) is a gap-filling and smoothing approach that mitigates these issues (Militino et al. 2019).
Filling missing data
Theoretical background
rsat implements a generic version of the algorithm with
the function smoothing_image().
nDays = 1 and
nYears = 1
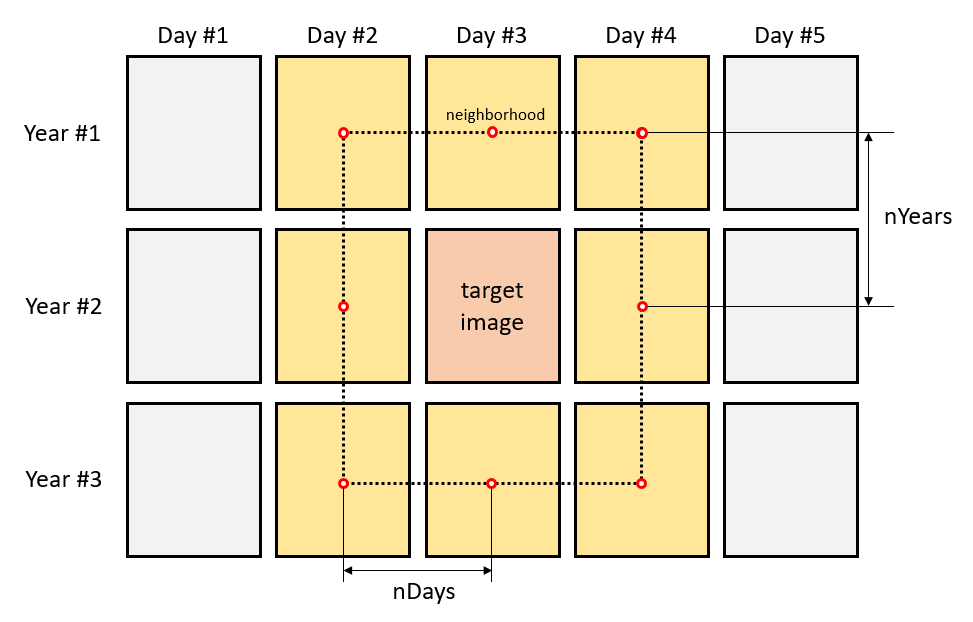
For the theoretical explanation, let’s assume that
images are available in the dataset (squares in the image above). The
imagery corresponds to
consecutive days over
years. Consider that we are willing to fill/smooth the target image (red
square). IMA fills the gaps borrowing information from an
adaptable temporal neighborhood (yellow squares). Two
parameters determine the size of the neighborhood; the number of days
before and after the target image (nDays) and the number of
previous and subsequent years (nYears). Both parameters
should be adjusted based on the temporal resolution of the of the
time-series of images. We recommend that the neighborhood extends over
days rather than years, when there is little resemblance between
seasons. Also, cloudy series may require larger neighborhoods.
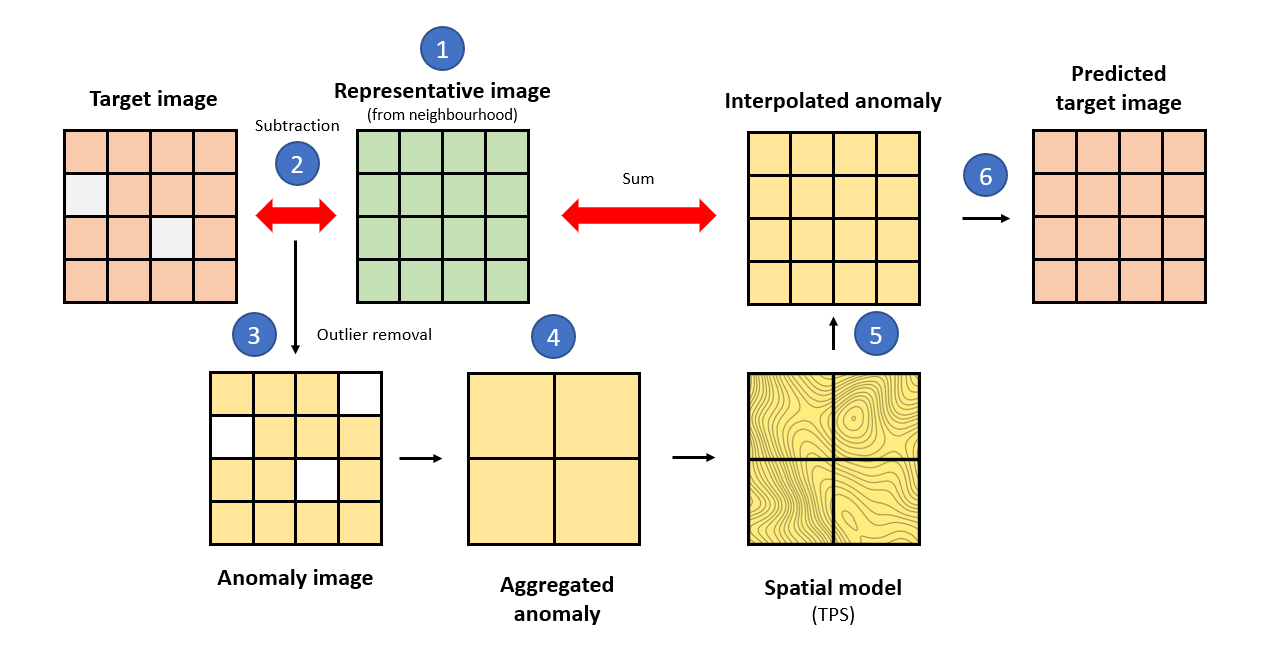
IMA gives the following steps; (1) creates a representative
image of the neighborhood ignoring missing values e.g., doing the mean,
median, etc. for each pixel’s time-series (fun), (2) the
target and representative images are subtracted giving an image of
anomalies, (3) the anomalies falling outside the
quantile limits (aFilter) are considered outliers and
therefore removed, (4) it aggregates the anomaly image into a coarser
resolution (fact) to reveal potential spatial dependencies,
(5) the procedure fits a spatial model (thin plate splines or
TPS) to the anomalies which is then used to interpolate the
values at the original resolution, and (6) the output is the sum of the
interpolated anomalies and the average image. The process is
encapsulated in smoothing_image() and the section below
shows its usage.
Hands-on demonstration
Data
The showcase uses a time-series of NDVI images. NDVI stands for Normalized Difference Vegetation Index and it is an indicator of vegetation’s vigor. Values close to indicate the presence of green and dense vegetation. Images correspond to a region in norther Spain between August and , in and . The images are part of the package and can be loaded into the environment as follows:
Let’s display the series of images to spot the gaps of data:
The maps show a hole on August , in the upper part of the image. The aim of IMA is to predict the values of the missing parts to provide a complete and continuous dataset.
Interpolation of Mean Anomalies
The instruction below applies the IMA to the entire series.
It uses a temporal neighborhood of
days and
year and the representative image is the mean. Outliers are
the anomalies outside
quantile range. Anomalies are aggregated at a factor of
(every
pixels are aggregated into
)
to fit an approximate spatial model. We recommend to tune the parameters
accordingly to obtain the best performance possible in each
situation:
library(rsat)
ndvi.fill <- rsat_smoothing_images(method = "IMA",
ex.ndvi.navarre,
nDays = 2,
nYears = 1,
fun = mean,
aFilter = c(0.01,0.99),
fact = 10,
only.na = TRUE)The algorithm predicts the value for the missing and the existing
pixels. The last argument, only.na = TRUE, indicates that
the results should preserve the original values wherever available.
IMA is able to run when neighborhoods are incomplete. In those
situations, the algorithm simply uses the imagery available within the
temporal neighborhood. The function prints a message specifying the
actual number of images being used in each case.
Let’s make a comparison between before and after the application of IMA.
before <- ex.ndvi.navarre[[1:3]]
after <- ndvi.fill[[1:3]]
tm_shape(c(before,after)) + tm_raster(title = "NDVI", style = "cont")The intention is to keep growing the number of algorithms devoted to processing in future versions of the package.
