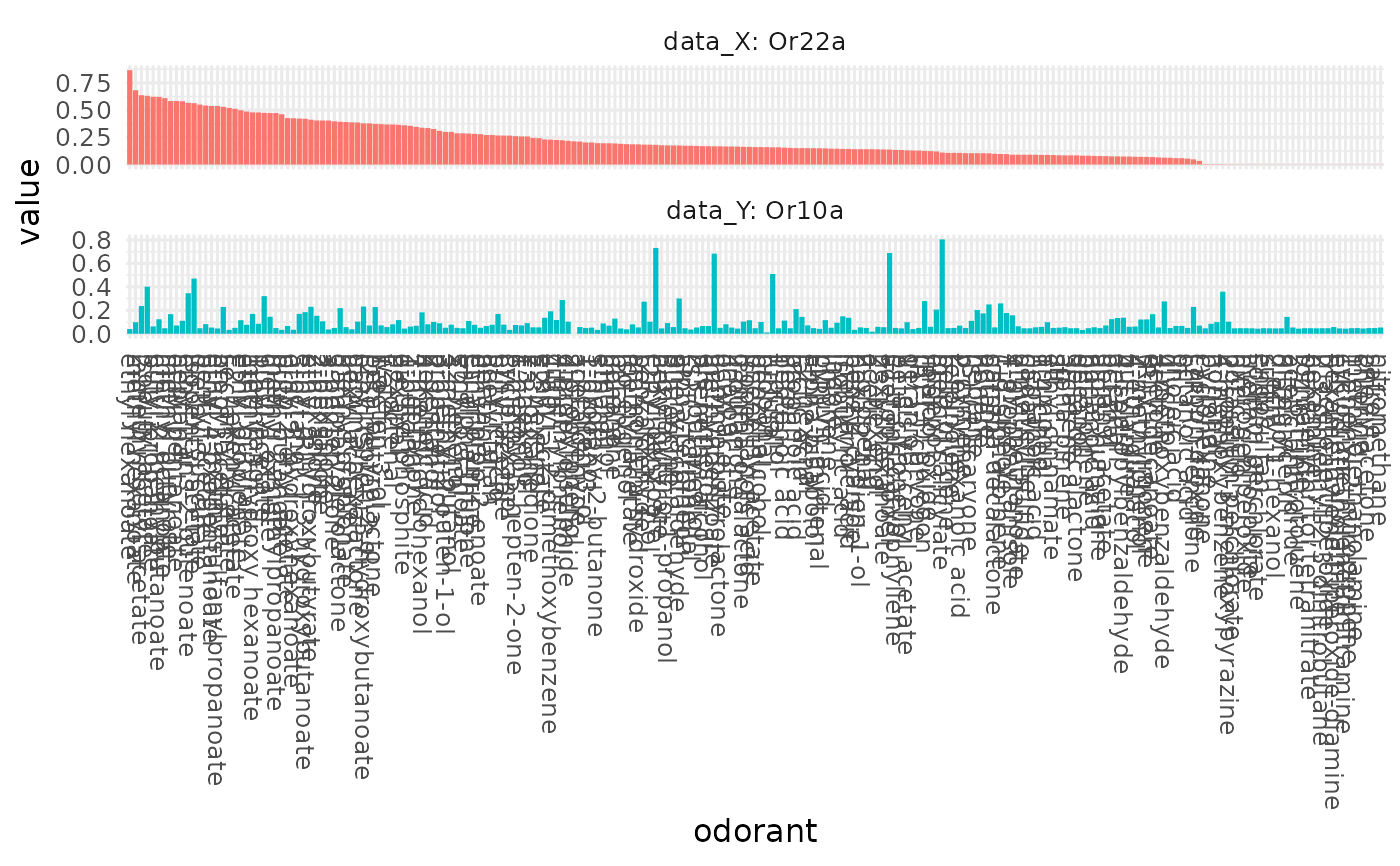
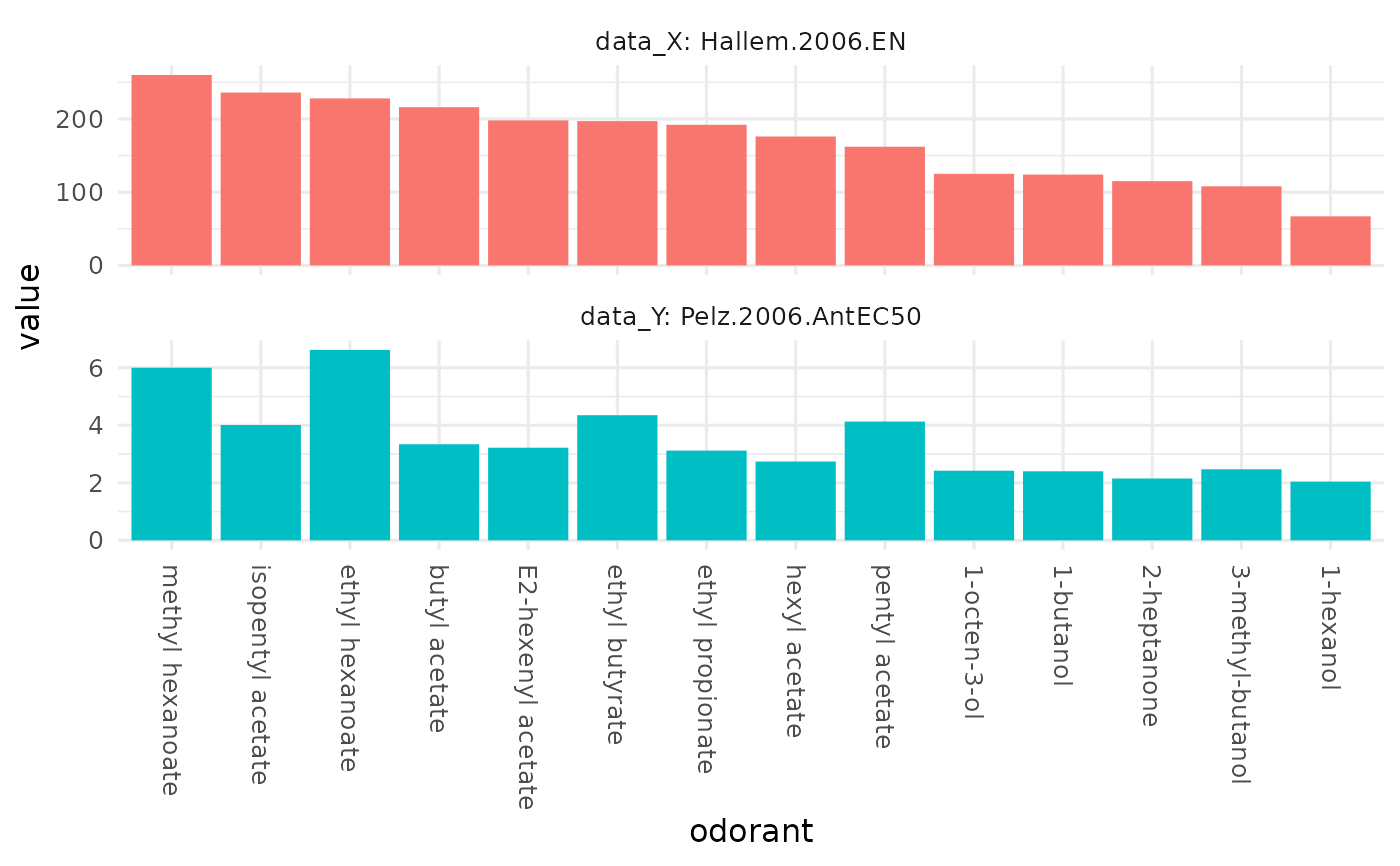
Orderdered bar plots for two studies, allowing for an easy comparison of the two studies / response profiles'.
Arguments
- x
the input data frame the first response profile will be taken from
- y
the input data frame the second response profile will be taken from (x will be taken if y is missing)
- by.x
character string, specifying a column in x
- by.y
character string, specifying a column in y
- tag
character, the chemical identifier that will be used as odorant label.
- base_size
numeric, the base font size for the ggplot2 plot
Author
Daniel Münch <daniel.muench@uni-konstanz.de>
Examples
# load data
library(DoOR.data)
library(DoOR.functions)
data(Or22a)
data(door_response_range)
data(door_response_matrix)
# compare the Hallem and the Pelz data set for Or22a
dplot_compare_profiles(x = Or22a, y = Or22a,
by.x = "Hallem.2006.EN",
by.y = "Pelz.2006.AntEC50")
 # comparedata from two different sensory neurons and add odorant labels
dplot_compare_profiles(x = cbind(door_response_matrix, InChIKey =
rownames(door_response_matrix)), y = cbind(door_response_matrix, InChIKey =
rownames(door_response_matrix)), by.x = "Or22a", by.y = "Or10a")
# comparedata from two different sensory neurons and add odorant labels
dplot_compare_profiles(x = cbind(door_response_matrix, InChIKey =
rownames(door_response_matrix)), y = cbind(door_response_matrix, InChIKey =
rownames(door_response_matrix)), by.x = "Or22a", by.y = "Or10a")