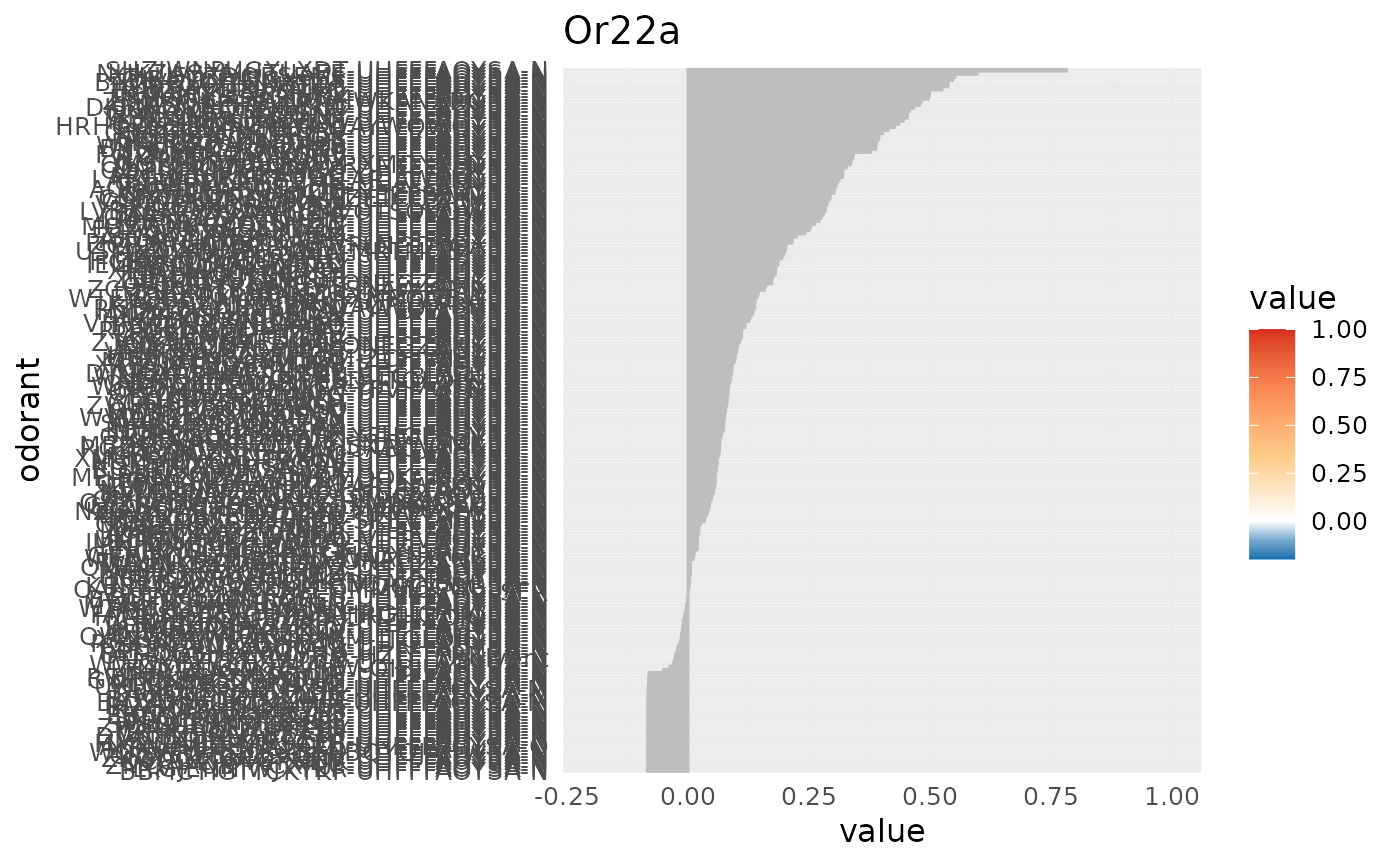
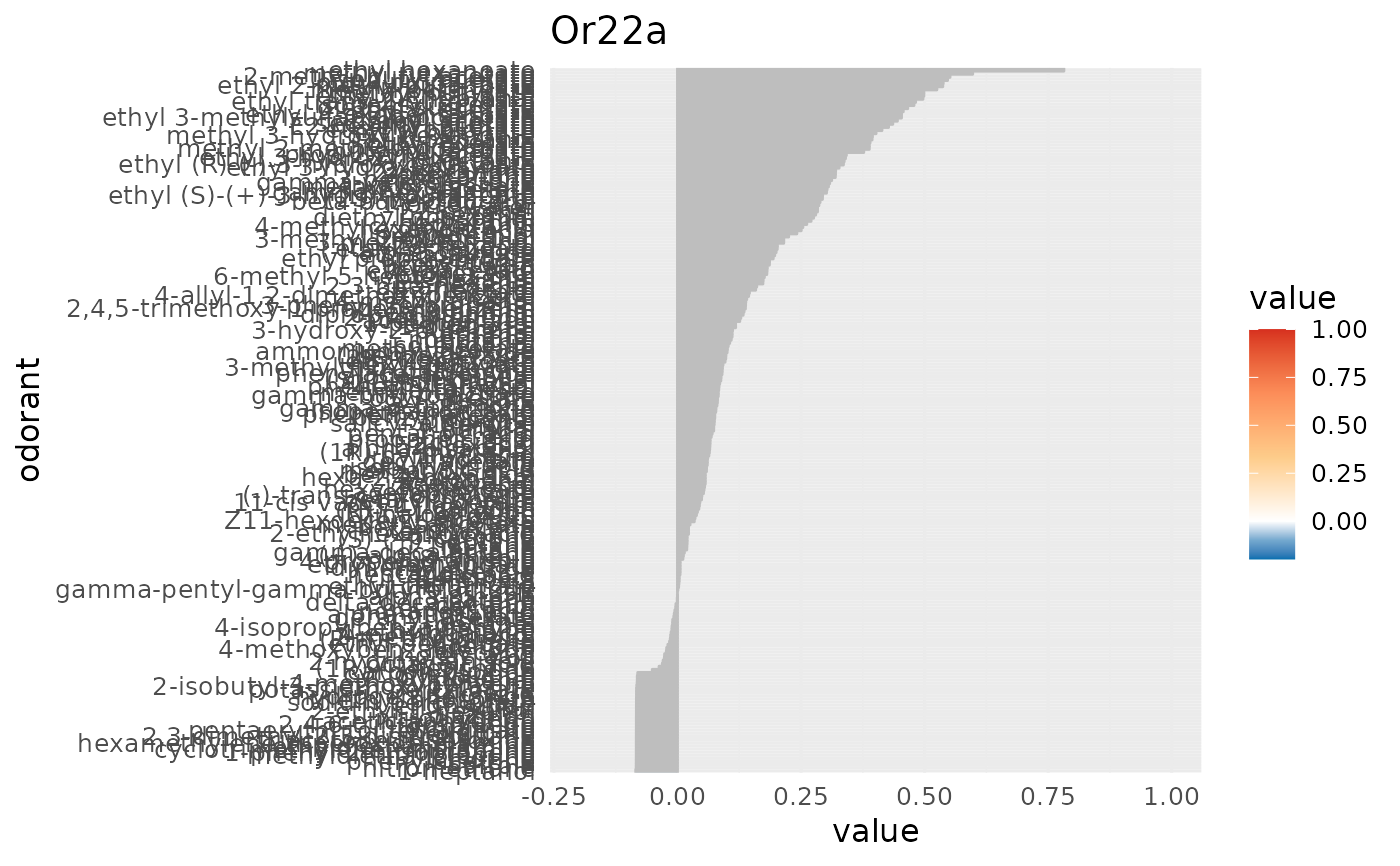
create a barplot of a DoOR response profile
Usage
dplot_response_profile(receptor,
response_matrix = door_default_values("door_response_matrix"),
odor_data = door_default_values("odor"), tag = door_default_values("tag"),
colored = TRUE, colors = door_default_values("colors"), limits,
zero = door_default_values("zero"),
scalebar = door_default_values("scalebar"), base_size = 12)Arguments
- receptor
character, receptor name, any of colnames(door_response_matrix)
- response_matrix
a DoOR door_response_matrix
- odor_data
data frame, contains the odorant information.
- tag
character, chemical identifier for annotation
- colored
logical, color code the bars according to the response value?
- colors
character vector, a vector of 5 colors (2 for values < 0, 1 value for 0 and 3 values > 0)
- limits
numeric of length 2, the limits for the colorscale and the x axis, global range of data will be used if empty
- zero
character, the odorant response that is set to 0, defaults to "SFR"
- scalebar
logical, add or suppress scalebars
- base_size
numeric, the base font size for the ggplot2 plot
Author
Daniel Münch <daniel.muench@uni-konstanz.de>