Introduction

The purpose of beautier is to create a valid
BEAST2 XML input file from its function argument. In this
way, a scientific pipeline using BEAST2 can be fully
scripted, instead of using BEAUti’s GUI.
beautier is part of the babette package
suite (website at https://github.com/ropensci/babette).
babette allows to use BEAST2 (and its tools) from R.
Getting started
For all examples, do load beautier:
Each example shows a picture of a BEAUti dialog to achieve the same.
BEAUti is part of the BEAST2 tool suite and it’s a GUI to create BEAST2
input files. beautier is an R package to supplement BEAUti,
by providing to do the same from an R script.
Each example reads the alignment from a FASTA file called
anthus_aco_sub.fas, which is part of the files supplied
with beautier:
input_filename <- get_beautier_path("anthus_aco_sub.fas")In this vignette, the generated BEAST2 XML is shown. Use
create_beast2_input_file to save the resulting XML directly
to file instead.
Example #1: all default
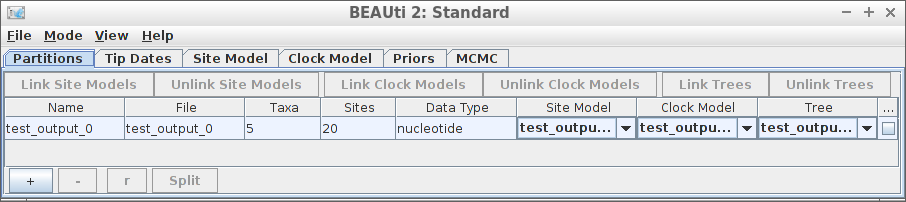
Using all default settings, only specify a DNA alignment.
create_beast2_input(
input_filename
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " </state>"
[43] ""
[44] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[45] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[46] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[47] " </populationModel>"
[48] " </init>"
[49] ""
[50] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[51] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[53] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[54] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[55] " </prior>"
[56] " </distribution>"
[57] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[58] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[59] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[60] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[61] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[62] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[63] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[64] " </siteModel>"
[65] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[66] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[67] " </branchRateModel>"
[68] " </distribution>"
[69] " </distribution>"
[70] " </distribution>"
[71] ""
[72] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[73] ""
[74] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[75] ""
[76] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[77] ""
[78] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[79] ""
[80] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[81] ""
[82] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[83] ""
[84] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[85] ""
[86] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[87] ""
[88] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[89] " <log idref=\"posterior\"/>"
[90] " <log idref=\"likelihood\"/>"
[91] " <log idref=\"prior\"/>"
[92] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[93] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[94] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[95] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[96] " </logger>"
[97] ""
[98] " <logger id=\"screenlog\" logEvery=\"1000\">"
[99] " <log idref=\"posterior\"/>"
[100] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[101] " <log idref=\"likelihood\"/>"
[102] " <log idref=\"prior\"/>"
[103] " </logger>"
[104] ""
[105] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[106] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[107] " </logger>"
[108] ""
[109] "</run>"
[110] ""
[111] "</beast>" All other parameters are set to their defaults, as in BEAUti.
Example #2: JC69 site model
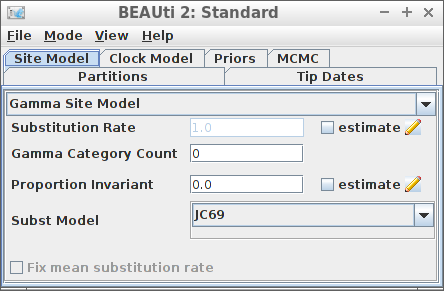
create_beast2_input(
input_filename,
site_model = create_jc69_site_model()
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " </state>"
[43] ""
[44] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[45] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[46] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[47] " </populationModel>"
[48] " </init>"
[49] ""
[50] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[51] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[53] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[54] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[55] " </prior>"
[56] " </distribution>"
[57] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[58] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[59] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[60] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[61] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[62] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[63] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[64] " </siteModel>"
[65] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[66] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[67] " </branchRateModel>"
[68] " </distribution>"
[69] " </distribution>"
[70] " </distribution>"
[71] ""
[72] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[73] ""
[74] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[75] ""
[76] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[77] ""
[78] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[79] ""
[80] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[81] ""
[82] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[83] ""
[84] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[85] ""
[86] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[87] ""
[88] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[89] " <log idref=\"posterior\"/>"
[90] " <log idref=\"likelihood\"/>"
[91] " <log idref=\"prior\"/>"
[92] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[93] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[94] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[95] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[96] " </logger>"
[97] ""
[98] " <logger id=\"screenlog\" logEvery=\"1000\">"
[99] " <log idref=\"posterior\"/>"
[100] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[101] " <log idref=\"likelihood\"/>"
[102] " <log idref=\"prior\"/>"
[103] " </logger>"
[104] ""
[105] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[106] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[107] " </logger>"
[108] ""
[109] "</run>"
[110] ""
[111] "</beast>" Example #3: Relaxed clock log normal
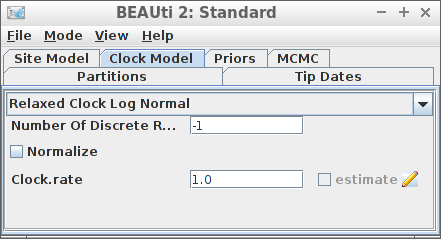
create_beast2_input(
input_filename,
clock_model = create_rln_clock_model()
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"ucldStdev.c:anthus_aco_sub\" lower=\"0.0\" name=\"stateNode\">0.1</parameter>"
[42] " <stateNode id=\"rateCategories.c:anthus_aco_sub\" spec=\"parameter.IntegerParameter\" dimension=\"8\">1</stateNode>"
[43] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[44] " </state>"
[45] ""
[46] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[47] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[48] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[49] " </populationModel>"
[50] " </init>"
[51] ""
[52] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[53] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[54] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[55] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[56] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[57] " </prior>"
[58] " <prior id=\"ucldStdevPrior.c:anthus_aco_sub\" name=\"distribution\" x=\"@ucldStdev.c:anthus_aco_sub\">"
[59] " <Gamma id=\"Gamma.0\" name=\"distr\">"
[60] " <parameter id=\"RealParameter.0\" estimate=\"false\" name=\"alpha\">0.5396</parameter>"
[61] " <parameter id=\"RealParameter.1\" estimate=\"false\" name=\"beta\">0.3819</parameter>"
[62] " </Gamma>"
[63] " </prior>"
[64] " </distribution>"
[65] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[66] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[67] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[68] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[69] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[70] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[71] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[72] " </siteModel>"
[73] " <branchRateModel id=\"RelaxedClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.UCRelaxedClockModel\" rateCategories=\"@rateCategories.c:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[74] " <LogNormal id=\"LogNormalDistributionModel.c:anthus_aco_sub\" S=\"@ucldStdev.c:anthus_aco_sub\" meanInRealSpace=\"true\" name=\"distr\">"
[75] " <parameter id=\"RealParameter.2\" estimate=\"false\" lower=\"0.0\" name=\"M\" upper=\"1.0\">1.0</parameter>"
[76] " </LogNormal>"
[77] " <parameter id=\"ucldMean.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[78] " </branchRateModel>"
[79] " </distribution>"
[80] " </distribution>"
[81] " </distribution>"
[82] ""
[83] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[84] ""
[85] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[86] ""
[87] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[88] ""
[89] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[90] ""
[91] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[92] ""
[93] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[94] ""
[95] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[96] ""
[97] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[98] ""
[99] " <operator id=\"ucldStdevScaler.c:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@ucldStdev.c:anthus_aco_sub\" scaleFactor=\"0.5\" weight=\"3.0\"/>"
[100] ""
[101] " <operator id=\"CategoriesRandomWalk.c:anthus_aco_sub\" spec=\"IntRandomWalkOperator\" parameter=\"@rateCategories.c:anthus_aco_sub\" weight=\"10.0\" windowSize=\"1\"/>"
[102] ""
[103] " <operator id=\"CategoriesSwapOperator.c:anthus_aco_sub\" spec=\"SwapOperator\" intparameter=\"@rateCategories.c:anthus_aco_sub\" weight=\"10.0\"/>"
[104] ""
[105] " <operator id=\"CategoriesUniform.c:anthus_aco_sub\" spec=\"UniformOperator\" parameter=\"@rateCategories.c:anthus_aco_sub\" weight=\"10.0\"/>"
[106] ""
[107] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[108] " <log idref=\"posterior\"/>"
[109] " <log idref=\"likelihood\"/>"
[110] " <log idref=\"prior\"/>"
[111] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[112] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[113] " <log idref=\"ucldStdev.c:anthus_aco_sub\"/>"
[114] " <log id=\"rate.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.RateStatistic\" branchratemodel=\"@RelaxedClock.c:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[115] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[116] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[117] " </logger>"
[118] ""
[119] " <logger id=\"screenlog\" logEvery=\"1000\">"
[120] " <log idref=\"posterior\"/>"
[121] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[122] " <log idref=\"likelihood\"/>"
[123] " <log idref=\"prior\"/>"
[124] " </logger>"
[125] ""
[126] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[127] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" branchratemodel=\"@RelaxedClock.c:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[128] " </logger>"
[129] ""
[130] "</run>"
[131] ""
[132] "</beast>" Example #4: Birth-Death tree prior
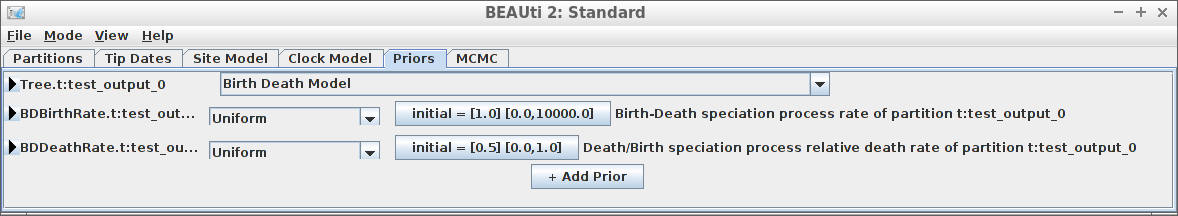
create_beast2_input(
input_filename,
tree_prior = create_bd_tree_prior()
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"BDBirthRate.t:anthus_aco_sub\" lower=\"0.0\" name=\"stateNode\" upper=\"10000.0\">1.0</parameter>"
[42] " <parameter id=\"BDDeathRate.t:anthus_aco_sub\" lower=\"0.0\" name=\"stateNode\" upper=\"1.0\">0.5</parameter>"
[43] " </state>"
[44] ""
[45] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[46] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[47] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[48] " </populationModel>"
[49] " </init>"
[50] ""
[51] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[53] " <distribution id=\"BirthDeath.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.BirthDeathGernhard08Model\" birthDiffRate=\"@BDBirthRate.t:anthus_aco_sub\" relativeDeathRate=\"@BDDeathRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[54] " <prior id=\"BirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@BDBirthRate.t:anthus_aco_sub\">"
[55] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[56] " </prior>"
[57] " <prior id=\"DeathRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@BDDeathRate.t:anthus_aco_sub\">"
[58] " <Uniform id=\"Uniform.101\" name=\"distr\" upper=\"Infinity\"/>"
[59] " </prior>"
[60] " </distribution>"
[61] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[62] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[63] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[64] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[65] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[66] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[67] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[68] " </siteModel>"
[69] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[70] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[71] " </branchRateModel>"
[72] " </distribution>"
[73] " </distribution>"
[74] " </distribution>"
[75] ""
[76] " <operator id=\"BirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@BDBirthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[77] ""
[78] " <operator id=\"DeathRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@BDDeathRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[79] ""
[80] " <operator id=\"BirthDeathTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[81] ""
[82] " <operator id=\"BirthDeathTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[83] ""
[84] " <operator id=\"BirthDeathUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[85] ""
[86] " <operator id=\"BirthDeathSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[87] ""
[88] " <operator id=\"BirthDeathNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[89] ""
[90] " <operator id=\"BirthDeathWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[91] ""
[92] " <operator id=\"BirthDeathWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[93] ""
[94] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[95] " <log idref=\"posterior\"/>"
[96] " <log idref=\"likelihood\"/>"
[97] " <log idref=\"prior\"/>"
[98] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[99] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[100] " <log idref=\"BirthDeath.t:anthus_aco_sub\"/>"
[101] " <log idref=\"BDBirthRate.t:anthus_aco_sub\"/>"
[102] " <log idref=\"BDDeathRate.t:anthus_aco_sub\"/>"
[103] " </logger>"
[104] ""
[105] " <logger id=\"screenlog\" logEvery=\"1000\">"
[106] " <log idref=\"posterior\"/>"
[107] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[108] " <log idref=\"likelihood\"/>"
[109] " <log idref=\"prior\"/>"
[110] " </logger>"
[111] ""
[112] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[113] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[114] " </logger>"
[115] ""
[116] "</run>"
[117] ""
[118] "</beast>" Example #5: Yule tree prior with a normally distributed birth rate
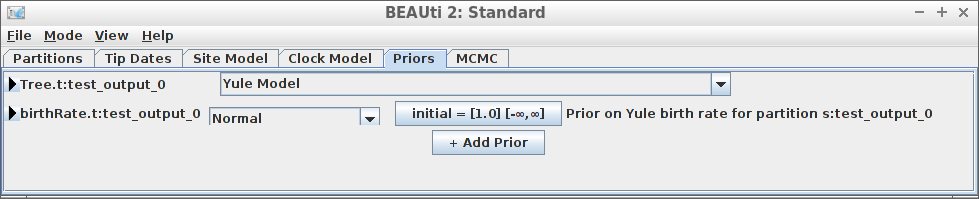
create_beast2_input(
input_filename,
tree_prior = create_yule_tree_prior(
birth_rate_distr = create_normal_distr()
)
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " </state>"
[43] ""
[44] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[45] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[46] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[47] " </populationModel>"
[48] " </init>"
[49] ""
[50] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[51] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[53] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[54] " <Normal id=\"Normal.100\" name=\"distr\">"
[55] " <parameter id=\"RealParameter.200\" estimate=\"false\" name=\"mean\">0</parameter>"
[56] " <parameter id=\"RealParameter.201\" estimate=\"false\" name=\"sigma\">1</parameter>"
[57] " </Normal>"
[58] " </prior>"
[59] " </distribution>"
[60] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[61] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[62] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[63] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[64] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[65] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[66] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[67] " </siteModel>"
[68] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[69] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[70] " </branchRateModel>"
[71] " </distribution>"
[72] " </distribution>"
[73] " </distribution>"
[74] ""
[75] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[76] ""
[77] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[78] ""
[79] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[80] ""
[81] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[82] ""
[83] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[84] ""
[85] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[86] ""
[87] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[88] ""
[89] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[90] ""
[91] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[92] " <log idref=\"posterior\"/>"
[93] " <log idref=\"likelihood\"/>"
[94] " <log idref=\"prior\"/>"
[95] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[96] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[97] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[98] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[99] " </logger>"
[100] ""
[101] " <logger id=\"screenlog\" logEvery=\"1000\">"
[102] " <log idref=\"posterior\"/>"
[103] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[104] " <log idref=\"likelihood\"/>"
[105] " <log idref=\"prior\"/>"
[106] " </logger>"
[107] ""
[108] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[109] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[110] " </logger>"
[111] ""
[112] "</run>"
[113] ""
[114] "</beast>" Thanks to Yacine Ben Chehida for this use case
Example #6: HKY site model with a non-zero proportion of invariants
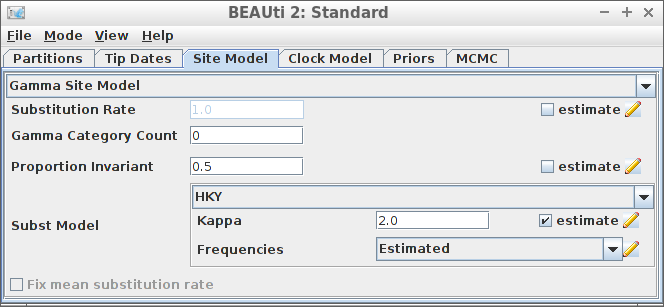
create_beast2_input(
input_filename,
site_model = create_hky_site_model(
gamma_site_model = create_gamma_site_model(prop_invariant = 0.5)
)
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"kappa.s:anthus_aco_sub\" lower=\"0.0\" name=\"stateNode\">2.0</parameter>"
[42] " <parameter id=\"freqParameter.s:anthus_aco_sub\" dimension=\"4\" lower=\"0.0\" name=\"stateNode\" upper=\"1.0\">0.25</parameter>"
[43] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[44] " </state>"
[45] ""
[46] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[47] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[48] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[49] " </populationModel>"
[50] " </init>"
[51] ""
[52] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[53] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[54] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[55] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[56] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[57] " </prior>"
[58] " <prior id=\"KappaPrior.s:anthus_aco_sub\" name=\"distribution\" x=\"@kappa.s:anthus_aco_sub\">"
[59] " <LogNormal id=\"LogNormalDistributionModel.0\" name=\"distr\">"
[60] " <parameter id=\"RealParameter.0\" estimate=\"false\" name=\"M\">1.0</parameter>"
[61] " <parameter id=\"RealParameter.1\" estimate=\"false\" name=\"S\">1.25</parameter>"
[62] " </LogNormal>"
[63] " </prior>"
[64] " </distribution>"
[65] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[66] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[67] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[68] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[69] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[70] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.5</parameter>"
[71] " <substModel id=\"hky.s:anthus_aco_sub\" spec=\"HKY\" kappa=\"@kappa.s:anthus_aco_sub\">"
[72] " <frequencies id=\"estimatedFreqs.s:anthus_aco_sub\" spec=\"Frequencies\" frequencies=\"@freqParameter.s:anthus_aco_sub\"/>"
[73] " </substModel>"
[74] " </siteModel>"
[75] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[76] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[77] " </branchRateModel>"
[78] " </distribution>"
[79] " </distribution>"
[80] " </distribution>"
[81] ""
[82] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[83] ""
[84] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[85] ""
[86] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[87] ""
[88] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[89] ""
[90] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[91] ""
[92] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[93] ""
[94] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[95] ""
[96] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[97] ""
[98] " <operator id=\"KappaScaler.s:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@kappa.s:anthus_aco_sub\" scaleFactor=\"0.5\" weight=\"0.1\"/>"
[99] ""
[100] " <operator id=\"FrequenciesExchanger.s:anthus_aco_sub\" spec=\"DeltaExchangeOperator\" delta=\"0.01\" weight=\"0.1\">"
[101] " <parameter idref=\"freqParameter.s:anthus_aco_sub\"/>"
[102] " </operator>"
[103] ""
[104] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[105] " <log idref=\"posterior\"/>"
[106] " <log idref=\"likelihood\"/>"
[107] " <log idref=\"prior\"/>"
[108] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[109] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[110] " <log idref=\"kappa.s:anthus_aco_sub\"/>"
[111] " <log idref=\"freqParameter.s:anthus_aco_sub\"/>"
[112] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[113] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[114] " </logger>"
[115] ""
[116] " <logger id=\"screenlog\" logEvery=\"1000\">"
[117] " <log idref=\"posterior\"/>"
[118] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[119] " <log idref=\"likelihood\"/>"
[120] " <log idref=\"prior\"/>"
[121] " </logger>"
[122] ""
[123] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[124] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[125] " </logger>"
[126] ""
[127] "</run>"
[128] ""
[129] "</beast>" Thanks to Yacine Ben Chehida for this use case
Example #7: Strict clock with a known clock rate
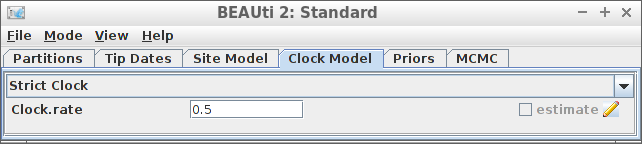
create_beast2_input(
input_filename,
clock_model = create_strict_clock_model(
clock_rate_param = 0.5
)
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " </state>"
[43] ""
[44] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[45] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[46] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[47] " </populationModel>"
[48] " </init>"
[49] ""
[50] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[51] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[53] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[54] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[55] " </prior>"
[56] " </distribution>"
[57] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[58] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[59] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[60] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[61] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[62] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[63] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[64] " </siteModel>"
[65] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[66] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">0.5</parameter>"
[67] " </branchRateModel>"
[68] " </distribution>"
[69] " </distribution>"
[70] " </distribution>"
[71] ""
[72] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[73] ""
[74] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[75] ""
[76] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[77] ""
[78] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[79] ""
[80] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[81] ""
[82] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[83] ""
[84] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[85] ""
[86] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[87] ""
[88] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[89] " <log idref=\"posterior\"/>"
[90] " <log idref=\"likelihood\"/>"
[91] " <log idref=\"prior\"/>"
[92] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[93] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[94] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[95] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[96] " </logger>"
[97] ""
[98] " <logger id=\"screenlog\" logEvery=\"1000\">"
[99] " <log idref=\"posterior\"/>"
[100] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[101] " <log idref=\"likelihood\"/>"
[102] " <log idref=\"prior\"/>"
[103] " </logger>"
[104] ""
[105] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[106] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[107] " </logger>"
[108] ""
[109] "</run>"
[110] ""
[111] "</beast>" Thanks to Paul van Els and Yacine Ben Chehida for this use case.
Example #8: Use MRCA prior
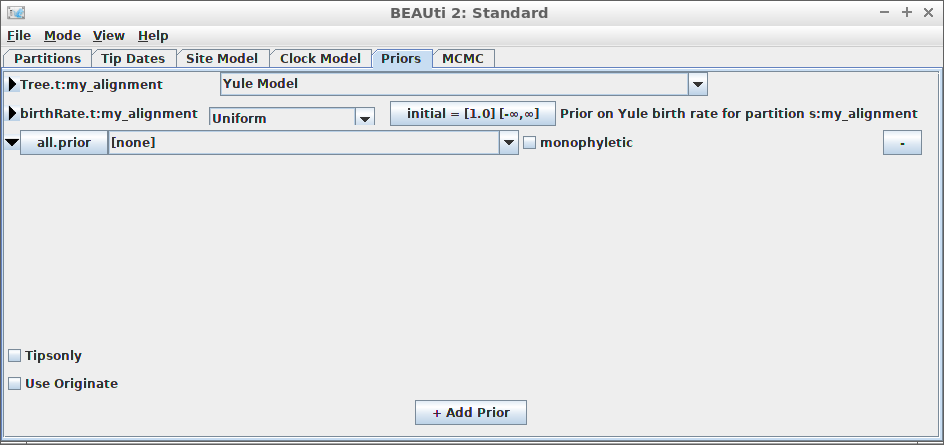
create_beast2_input(
input_filename,
mrca_prior = create_mrca_prior()
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " </state>"
[43] ""
[44] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[45] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[46] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[47] " </populationModel>"
[48] " </init>"
[49] ""
[50] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[51] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[53] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[54] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[55] " </prior>"
[56] " <distribution id=\"auto_name_1.prior\" spec=\"beast.math.distributions.MRCAPrior\" tree=\"@Tree.t:anthus_aco_sub\">"
[57] " <taxonset id=\"auto_name_1\" spec=\"TaxonSet\">"
[58] " <taxon id=\"61430_aco\" spec=\"Taxon\"/>"
[59] " <taxon id=\"626029_aco\" spec=\"Taxon\"/>"
[60] " <taxon id=\"630116_aco\" spec=\"Taxon\"/>"
[61] " <taxon id=\"630210_aco\" spec=\"Taxon\"/>"
[62] " <taxon id=\"B25702_aco\" spec=\"Taxon\"/>"
[63] " </taxonset>"
[64] " </distribution>"
[65] " </distribution>"
[66] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[67] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[68] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[69] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[70] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[71] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[72] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[73] " </siteModel>"
[74] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\">"
[75] " <parameter id=\"clockRate.c:anthus_aco_sub\" estimate=\"false\" name=\"clock.rate\">1.0</parameter>"
[76] " </branchRateModel>"
[77] " </distribution>"
[78] " </distribution>"
[79] " </distribution>"
[80] ""
[81] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[82] ""
[83] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[84] ""
[85] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[86] ""
[87] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[88] ""
[89] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[90] ""
[91] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[92] ""
[93] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[94] ""
[95] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[96] ""
[97] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[98] " <log idref=\"posterior\"/>"
[99] " <log idref=\"likelihood\"/>"
[100] " <log idref=\"prior\"/>"
[101] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[102] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[103] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[104] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[105] " <log idref=\"auto_name_1.prior\"/>"
[106] " </logger>"
[107] ""
[108] " <logger id=\"screenlog\" logEvery=\"1000\">"
[109] " <log idref=\"posterior\"/>"
[110] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[111] " <log idref=\"likelihood\"/>"
[112] " <log idref=\"prior\"/>"
[113] " </logger>"
[114] ""
[115] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[116] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[117] " </logger>"
[118] ""
[119] "</run>"
[120] ""
[121] "</beast>" Example #9: Use MRCA prior to specify a close-to-fixed crown age
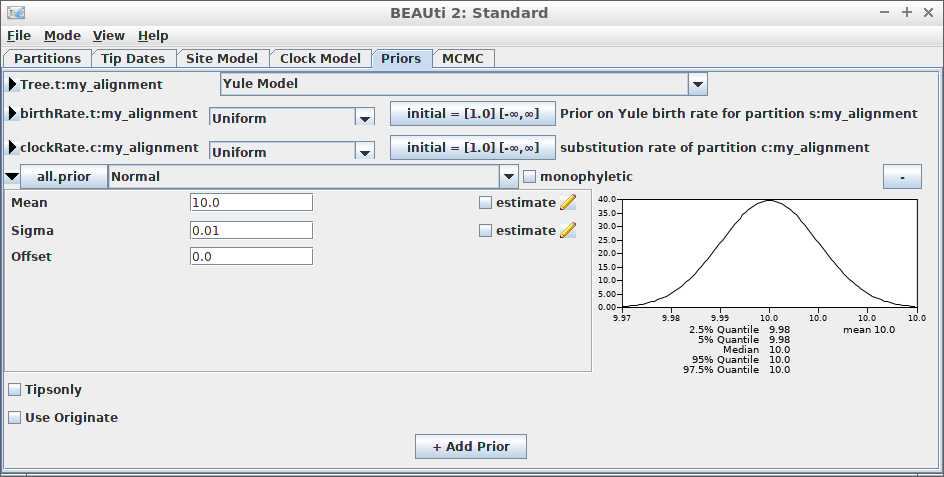
With an MRCA prior, it is possible to specify a close-to-fixed crown age:
crown_age <- 15
create_beast2_input(
input_filename,
mrca_prior = create_mrca_prior(
is_monophyletic = TRUE,
mrca_distr = create_normal_distr(
mean = crown_age,
sigma = 0.001
)
)
)
[1] "<?xml version=\"1.0\" encoding=\"UTF-8\" standalone=\"no\"?><beast beautitemplate='Standard' beautistatus='' namespace=\"beast.core:beast.evolution.alignment:beast.evolution.tree.coalescent:beast.core.util:beast.evolution.nuc:beast.evolution.operators:beast.evolution.sitemodel:beast.evolution.substitutionmodel:beast.evolution.likelihood\" required=\"\" version=\"2.4\">"
[2] ""
[3] ""
[4] " <data"
[5] "id=\"anthus_aco_sub\""
[6] "name=\"alignment\">"
[7] " <sequence id=\"seq_61430_aco\" taxon=\"61430_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[8] " <sequence id=\"seq_626029_aco\" taxon=\"626029_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctgcttgtttaatgccctctcctattttattgtgacgattgtctgttttt\"/>"
[9] " <sequence id=\"seq_630116_aco\" taxon=\"630116_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgcgaaaattgtctgttttt\"/>"
[10] " <sequence id=\"seq_630210_aco\" taxon=\"630210_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[11] " <sequence id=\"seq_B25702_aco\" taxon=\"B25702_aco\" totalcount=\"4\" value=\"acaggttagaaactactctgttttctggctccttgtttaatgccctgtcctattttattgtgacaattgtctgttttt\"/>"
[12] " </data>"
[13] ""
[14] ""
[15] " "
[16] ""
[17] ""
[18] " "
[19] ""
[20] ""
[21] " "
[22] "<map name=\"Uniform\" >beast.math.distributions.Uniform</map>"
[23] "<map name=\"Exponential\" >beast.math.distributions.Exponential</map>"
[24] "<map name=\"LogNormal\" >beast.math.distributions.LogNormalDistributionModel</map>"
[25] "<map name=\"Normal\" >beast.math.distributions.Normal</map>"
[26] "<map name=\"Beta\" >beast.math.distributions.Beta</map>"
[27] "<map name=\"Gamma\" >beast.math.distributions.Gamma</map>"
[28] "<map name=\"LaplaceDistribution\" >beast.math.distributions.LaplaceDistribution</map>"
[29] "<map name=\"prior\" >beast.math.distributions.Prior</map>"
[30] "<map name=\"InverseGamma\" >beast.math.distributions.InverseGamma</map>"
[31] "<map name=\"OneOnX\" >beast.math.distributions.OneOnX</map>"
[32] ""
[33] ""
[34] "<run id=\"mcmc\" spec=\"MCMC\" chainLength=\"10000000\">"
[35] " <state id=\"state\" storeEvery=\"5000\">"
[36] " <tree id=\"Tree.t:anthus_aco_sub\" name=\"stateNode\">"
[37] " <taxonset id=\"TaxonSet.anthus_aco_sub\" spec=\"TaxonSet\">"
[38] " <alignment idref=\"anthus_aco_sub\"/>"
[39] " </taxonset>"
[40] " </tree>"
[41] " <parameter id=\"birthRate.t:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[42] " <parameter id=\"clockRate.c:anthus_aco_sub\" name=\"stateNode\">1.0</parameter>"
[43] " </state>"
[44] ""
[45] " <init id=\"RandomTree.t:anthus_aco_sub\" spec=\"beast.evolution.tree.RandomTree\" estimate=\"false\" initial=\"@Tree.t:anthus_aco_sub\" taxa=\"@anthus_aco_sub\">"
[46] " <populationModel id=\"ConstantPopulation0.t:anthus_aco_sub\" spec=\"ConstantPopulation\">"
[47] " <parameter id=\"randomPopSize.t:anthus_aco_sub\" name=\"popSize\">1.0</parameter>"
[48] " </populationModel>"
[49] " </init>"
[50] ""
[51] " <distribution id=\"posterior\" spec=\"util.CompoundDistribution\">"
[52] " <distribution id=\"prior\" spec=\"util.CompoundDistribution\">"
[53] " <distribution id=\"YuleModel.t:anthus_aco_sub\" spec=\"beast.evolution.speciation.YuleModel\" birthDiffRate=\"@birthRate.t:anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[54] " <prior id=\"YuleBirthRatePrior.t:anthus_aco_sub\" name=\"distribution\" x=\"@birthRate.t:anthus_aco_sub\">"
[55] " <Uniform id=\"Uniform.100\" name=\"distr\" upper=\"Infinity\"/>"
[56] " </prior>"
[57] " <prior id=\"ClockPrior.c:anthus_aco_sub\" name=\"distribution\" x=\"@clockRate.c:anthus_aco_sub\">"
[58] " <Uniform id=\"Uniform.150\" name=\"distr\" upper=\"Infinity\"/>"
[59] " </prior>"
[60] " <distribution id=\"auto_name_1.prior\" spec=\"beast.math.distributions.MRCAPrior\" monophyletic=\"true\" tree=\"@Tree.t:anthus_aco_sub\">"
[61] " <taxonset id=\"auto_name_1\" spec=\"TaxonSet\">"
[62] " <taxon id=\"61430_aco\" spec=\"Taxon\"/>"
[63] " <taxon id=\"626029_aco\" spec=\"Taxon\"/>"
[64] " <taxon id=\"630116_aco\" spec=\"Taxon\"/>"
[65] " <taxon id=\"630210_aco\" spec=\"Taxon\"/>"
[66] " <taxon id=\"B25702_aco\" spec=\"Taxon\"/>"
[67] " </taxonset>"
[68] " <Normal id=\"Normal.151\" name=\"distr\">"
[69] " <parameter id=\"RealParameter.300\" estimate=\"false\" name=\"mean\">15</parameter>"
[70] " <parameter id=\"RealParameter.301\" estimate=\"false\" name=\"sigma\">0.001</parameter>"
[71] " </Normal>"
[72] " </distribution>"
[73] " </distribution>"
[74] " <distribution id=\"likelihood\" spec=\"util.CompoundDistribution\" useThreads=\"true\">"
[75] " <distribution id=\"treeLikelihood.anthus_aco_sub\" spec=\"ThreadedTreeLikelihood\" data=\"@anthus_aco_sub\" tree=\"@Tree.t:anthus_aco_sub\">"
[76] " <siteModel id=\"SiteModel.s:anthus_aco_sub\" spec=\"SiteModel\">"
[77] " <parameter id=\"mutationRate.s:anthus_aco_sub\" estimate=\"false\" name=\"mutationRate\">1.0</parameter>"
[78] " <parameter id=\"gammaShape.s:anthus_aco_sub\" estimate=\"false\" name=\"shape\">1.0</parameter>"
[79] " <parameter id=\"proportionInvariant.s:anthus_aco_sub\" estimate=\"false\" lower=\"0.0\" name=\"proportionInvariant\" upper=\"1.0\">0.0</parameter>"
[80] " <substModel id=\"JC69.s:anthus_aco_sub\" spec=\"JukesCantor\"/>"
[81] " </siteModel>"
[82] " <branchRateModel id=\"StrictClock.c:anthus_aco_sub\" spec=\"beast.evolution.branchratemodel.StrictClockModel\" clock.rate=\"@clockRate.c:anthus_aco_sub\"/>"
[83] " </distribution>"
[84] " </distribution>"
[85] " </distribution>"
[86] ""
[87] " <operator id=\"YuleBirthRateScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@birthRate.t:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[88] ""
[89] " <operator id=\"YuleModelTreeScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[90] ""
[91] " <operator id=\"YuleModelTreeRootScaler.t:anthus_aco_sub\" spec=\"ScaleOperator\" rootOnly=\"true\" scaleFactor=\"0.5\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[92] ""
[93] " <operator id=\"YuleModelUniformOperator.t:anthus_aco_sub\" spec=\"Uniform\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"30.0\"/>"
[94] ""
[95] " <operator id=\"YuleModelSubtreeSlide.t:anthus_aco_sub\" spec=\"SubtreeSlide\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[96] ""
[97] " <operator id=\"YuleModelNarrow.t:anthus_aco_sub\" spec=\"Exchange\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"15.0\"/>"
[98] ""
[99] " <operator id=\"YuleModelWide.t:anthus_aco_sub\" spec=\"Exchange\" isNarrow=\"false\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[100] ""
[101] " <operator id=\"YuleModelWilsonBalding.t:anthus_aco_sub\" spec=\"WilsonBalding\" tree=\"@Tree.t:anthus_aco_sub\" weight=\"3.0\"/>"
[102] ""
[103] " <operator id=\"StrictClockRateScaler.c:anthus_aco_sub\" spec=\"ScaleOperator\" parameter=\"@clockRate.c:anthus_aco_sub\" scaleFactor=\"0.75\" weight=\"3.0\"/>"
[104] ""
[105] " <operator id=\"strictClockUpDownOperator.c:anthus_aco_sub\" spec=\"UpDownOperator\" scaleFactor=\"0.75\" weight=\"3.0\">"
[106] " <up idref=\"clockRate.c:anthus_aco_sub\"/>"
[107] " <down idref=\"Tree.t:anthus_aco_sub\"/>"
[108] " </operator>"
[109] ""
[110] " <logger id=\"tracelog\" fileName=\"anthus_aco_sub.log\" logEvery=\"1000\" model=\"@posterior\" sanitiseHeaders=\"true\" sort=\"smart\">"
[111] " <log idref=\"posterior\"/>"
[112] " <log idref=\"likelihood\"/>"
[113] " <log idref=\"prior\"/>"
[114] " <log idref=\"treeLikelihood.anthus_aco_sub\"/>"
[115] " <log id=\"TreeHeight.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeHeightLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[116] " <log idref=\"YuleModel.t:anthus_aco_sub\"/>"
[117] " <log idref=\"birthRate.t:anthus_aco_sub\"/>"
[118] " <log idref=\"auto_name_1.prior\"/>"
[119] " <log idref=\"clockRate.c:anthus_aco_sub\"/>"
[120] " </logger>"
[121] ""
[122] " <logger id=\"screenlog\" logEvery=\"1000\">"
[123] " <log idref=\"posterior\"/>"
[124] " <log id=\"ESS.0\" spec=\"util.ESS\" arg=\"@posterior\"/>"
[125] " <log idref=\"likelihood\"/>"
[126] " <log idref=\"prior\"/>"
[127] " </logger>"
[128] ""
[129] " <logger id=\"treelog.t:anthus_aco_sub\" fileName=\"$(tree).trees\" logEvery=\"1000\" mode=\"tree\">"
[130] " <log id=\"TreeWithMetaDataLogger.t:anthus_aco_sub\" spec=\"beast.evolution.tree.TreeWithMetaDataLogger\" tree=\"@Tree.t:anthus_aco_sub\"/>"
[131] " </logger>"
[132] ""
[133] "</run>"
[134] ""
[135] "</beast>" Cleanup
beautier::remove_beautier_folder()
beautier::check_empty_beautier_folder()