Bayesian data analysis usually incurs long runtimes and cumbersome custom code, and the process of prototyping and deploying custom JAGS models can become a daunting software engineering challenge. To ease this burden, the jagstargets R package creates JAGS pipelines that are concise, efficient, scalable, and tailored to the needs of Bayesian statisticians. Leveraging targets, jagstargets pipelines automatically parallelize the computation and skip expensive steps when the results are already up to date. Minimal custom user-side code is required, and there is no need to manually configure branching, so jagstargets is easier to use than targets and R2jags directly.
Prerequisites
- The prerequisites of the
targetsR package. - Basic familiarity with
targets: watch minutes 7 through 40 of this video, then read this chapter of the user manual. - Familiarity with Bayesian Statistics and JAGS. Prior knowledge of
rjagsorR2jagshelps.
How to get started
Read the jagstargets introductory vignette, and then use https://docs.ropensci.org/jagstargets/ as a reference while constructing your own workflows. If you need to analyze large collections of simulated datasets, please consult the simulation vignette.
Installation
jagstargets requires the user to install JAGS, rjags, and R2jags beforehand. You can install JAGS from https://mcmc-jags.sourceforge.io/, and you can install the rest from CRAN.
install.packages(c("rjags", "R2jags"))Then, install the latest release from CRAN.
install.packages("jagstargets")Alternatively, install the GitHub development version to access the latest features and patches.
install.packages("remotes")
remotes::install_github("ropensci/jagstargets")Usage
Begin with one or more models: for example, the simple regression model below with response variable \(y\) and covariate \(x\).
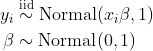
Next, write a JAGS model file for each model like the model.jags file below.
model {
for (i in 1:n) {
y[i] ~ dnorm(x[i] * beta, 1)
}
beta ~ dnorm(0, 1)
}To begin a reproducible analysis pipeline with this model, write a _targets.R file that loads your packages, defines a function to generate JAGS data, and lists a pipeline of targets. The target list can call target factories like tar_jags() as well as ordinary targets with tar_target(). The following minimal example is simple enough to contain entirely within the _targets.R file, but for larger projects, you may wish to store functions in separate files as in the targets-stan example.
# _targets.R
library(targets)
library(jagstargets)
generate_data <- function() {
true_beta <- stats::rnorm(n = 1, mean = 0, sd = 1)
x <- seq(from = -1, to = 1, length.out = n)
y <- stats::rnorm(n, x * true_beta, 1)
out <- list(n = n, x = x, y = y, true_beta = true_beta)
}
list(
tar_jags(
example,
jags_files = "model.jags", # You provide this file.
parameters.to.save = "beta",
data = generate_data()
)
)Run tar_visnetwork() to check _targets.R for correctness, then call tar_make() to run the pipeline. Access the results using tar_read(), e.g. tar_read(tar_read(example_summary_x). Visit the introductory vignette to read more about this example.
How the package works
jagstargets supports specialized target factories that create ensembles of target objects for R2jags workflows. These target factories abstract away the details of targets and R2jags and make both packages easier to use. For details, please read the introductory vignette.
Help
Please read the targets help guide at https://books.ropensci.org/targets/help.html to learn how to ask for help.
If you have trouble using jagstargets, you can ask for help in the GitHub discussions forum. Because the purpose of jagstargets is to combine targets and R2jags, your issue may have something to do with one of the latter two packages, a dependency of targets, or R2jags itself. When you troubleshoot, peel back as many layers as possible to isolate the problem. For example, if the issue comes from R2jags, create a reproducible example that directly invokes R2jags without invoking jagstargets. The GitHub discussion and issue forums of those packages are great resources.
Participation
Development is a community effort, and we welcome discussion and contribution. By participating in this project, you agree to abide by the code of conduct and the contributing guide.
Citation
citation("jagstargets")
#>
#> To cite jagstargets in publications use:
#>
#> Landau, W. M., (2021). The jagstargets R package: a reproducible
#> workflow framework for Bayesian data analysis with JAGS. Journal of
#> Open Source Software, 6(68), 3877,
#> https://doi.org/10.21105/joss.03877
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {The jagstargets R package: a reproducible workflow framework for Bayesian data analysis with JAGS},
#> author = {William Michael Landau},
#> journal = {Journal of Open Source Software},
#> year = {2021},
#> volume = {6},
#> number = {68},
#> pages = {3877},
#> url = {https://doi.org/10.21105/joss.03877},
#> }