
Simple search and citation of occurrences
Hannah L. Owens
Cory Merow
Brian Maitner
Jamie M. Kass
Vijay Barve
Robert Guralnick
2026-02-15
Source:vignettes/a_Simple.Rmd
a_Simple.RmdIntroduction
We have entered the age of data-intensive scientific discovery. As
data sets increase in complexity and heterogeneity, we must preserve the
cycle of data citation from primary data sources to aggregating
databases to research products and back to primary data sources. The
citation cycle keeps science transparent, but it is also key to
supporting primary providers by documenting the use of their data. The
Global Biodiversity Information Facility (GBIF), Botanical Information
and Ecology Network (BIEN), and other data aggregators have made great
strides in harvesting citation data from research products and linking
them back to primary data providers. However, this only works if those
that publish research products cite primary data sources in the first
place. We developed occCite, a set of R-based
tools for downloading, managing, and citing biodiversity data, to
advance toward the goal of closing the data provenance cycle. These
tools preserve links between occurrence data and primary providers once
researchers download aggregated data, and facilitate the citation of
primary data providers in research papers.
The occCite workflow follows a three-step process.
First, the user inputs one or more taxonomic names (or a phylogeny).
occCite then rectifies these names by checking them against
one or more taxonomic databases, which can be specified by the user (see
the Global Names
List). The results of the taxonomic rectification are then kept in
an occCiteData object in local memory. Next,
occCite takes the occCiteData object and
user-defined search parameters to query BIEN (through
rbien) and/or GBIF(through rGBIF) for records.
The results are appended to the occCiteData object, along
with metadata on the search. Finally, the user can pass the
occCiteData object to occCitation, which
compiles citations for the primary providers, database aggregators, and
R packages used to build the dataset.
Future iterations of occCite will track citation data
through the data cleaning process and provide a series of visualizations
on raw query results and final data sets. It will also provide data
citations in a format congruent with best-practice recommendations for
large biodiversity data sets. Based on these data citation tools, we
will also propose a new set of standards for citing primary biodiversity
data in published research articles that provides due credit to
contributors and allows them to track the use of their work. Keep
checking back!
Setup
If you plan to query GBIF, you will need to provide them with your
user login information. We have provided a dummy login below to show you
the format. You will need to provide actual account
information. This is because you will actually be downloading
all of the records available for the species using
occ_download(), instead of getting results from
occ_search(), which has a hard limit of 100,000
occurrences.
library(occCite);
#Creating a GBIF login
GBIFLogin <- GBIFLoginManager(user = "occCiteTester",
email = "****@yahoo.com",
pwd = "12345")Performing a simple search
The basics
At its simplest, occCite allows you to search for
occurrences for a single species. The taxonomy of the user-specified
species will be verified using EOL and NCBI taxonomies by default.
# Simple search
mySimpleOccCiteObject <- occQuery(x = "Protea cynaroides",
datasources = c("gbif", "bien"),
GBIFLogin = GBIFLogin,
GBIFDownloadDirectory =
system.file('extdata/', package='occCite'),
checkPreviousGBIFDownload = T)Here is what the GBIF results look like:
# GBIF search results
head(mySimpleOccCiteObject@occResults$`Protea cynaroides`$GBIF$OccurrenceTable)## name longitude latitude coordinateUncertaintyInMeters day month
## 1 Protea cynaroides 18.43928 -33.95440 8 17 2
## 2 Protea cynaroides 22.12754 -33.91561 4 11 2
## 3 Protea cynaroides 18.43927 -33.95429 8 17 2
## 4 Protea cynaroides 18.43254 -34.29275 31 6 2
## 5 Protea cynaroides 18.42429 -34.02934 2167 10 2
## 6 Protea cynaroides 18.43529 -34.10545 2 8 2
## year datasetKey dataService
## 1 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 2 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 3 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 4 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 5 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 6 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## datasetName
## 1 iNaturalist Research-grade Observations
## 2 iNaturalist Research-grade Observations
## 3 iNaturalist Research-grade Observations
## 4 iNaturalist Research-grade Observations
## 5 iNaturalist Research-grade Observations
## 6 iNaturalist Research-grade ObservationsAnd here are the BIEN results:
#BIEN search results
head(mySimpleOccCiteObject@occResults$`Protea cynaroides`$BIEN$OccurrenceTable)## name longitude latitude coordinateUncertaintyInMeters day month
## 1 Protea cynaroides 19.14767 -33.7137 NA 30 1
## 2 Protea cynaroides 18.62500 -32.6250 NA 24 10
## 3 Protea cynaroides 19.12500 -34.3750 NA 22 7
## 4 Protea cynaroides 19.62500 -34.6250 NA 10 4
## 5 Protea cynaroides 19.37500 -33.1250 NA 30 3
## 6 Protea cynaroides 19.37500 -34.1250 NA 12 3
## year datasetName datasetKey dataService
## 1 1828 MNHN 4620 BIEN
## 2 1954 SANBI 3318 BIEN
## 3 1967 SANBI 3318 BIEN
## 4 1979 SANBI 3318 BIEN
## 5 1978 SANBI 3318 BIEN
## 6 1962 SANBI 3318 BIENThere is also a summary method for occCite objects with
some basic information about your search.
summary(mySimpleOccCiteObject)##
## OccCite query occurred on: 20 June, 2024
##
## User query type: User-supplied list of taxa.
##
## Sources for taxonomic rectification: GBIF Backbone Taxonomy
##
##
## Taxonomic cleaning results:
##
## Input Name Best Match Taxonomic Databases w/ Matches
## 1 Protea cynaroides Protea cynaroides (L.) L. GBIF Backbone Taxonomy
##
## Sources for occurrence data: gbif, bien
##
## Species Occurrences Sources
## 1 Protea cynaroides (L.) L. 2334 17
##
## GBIF dataset DOIs:
##
## Species GBIF Access Date GBIF DOI
## 1 Protea cynaroides (L.) L. 2022-03-02 10.15468/dl.ztbx8cIf you want to visualize the results of your search, you can use the
plot method on occCite objects to generate
several kinds of summary plots.
plot(mySimpleOccCiteObject)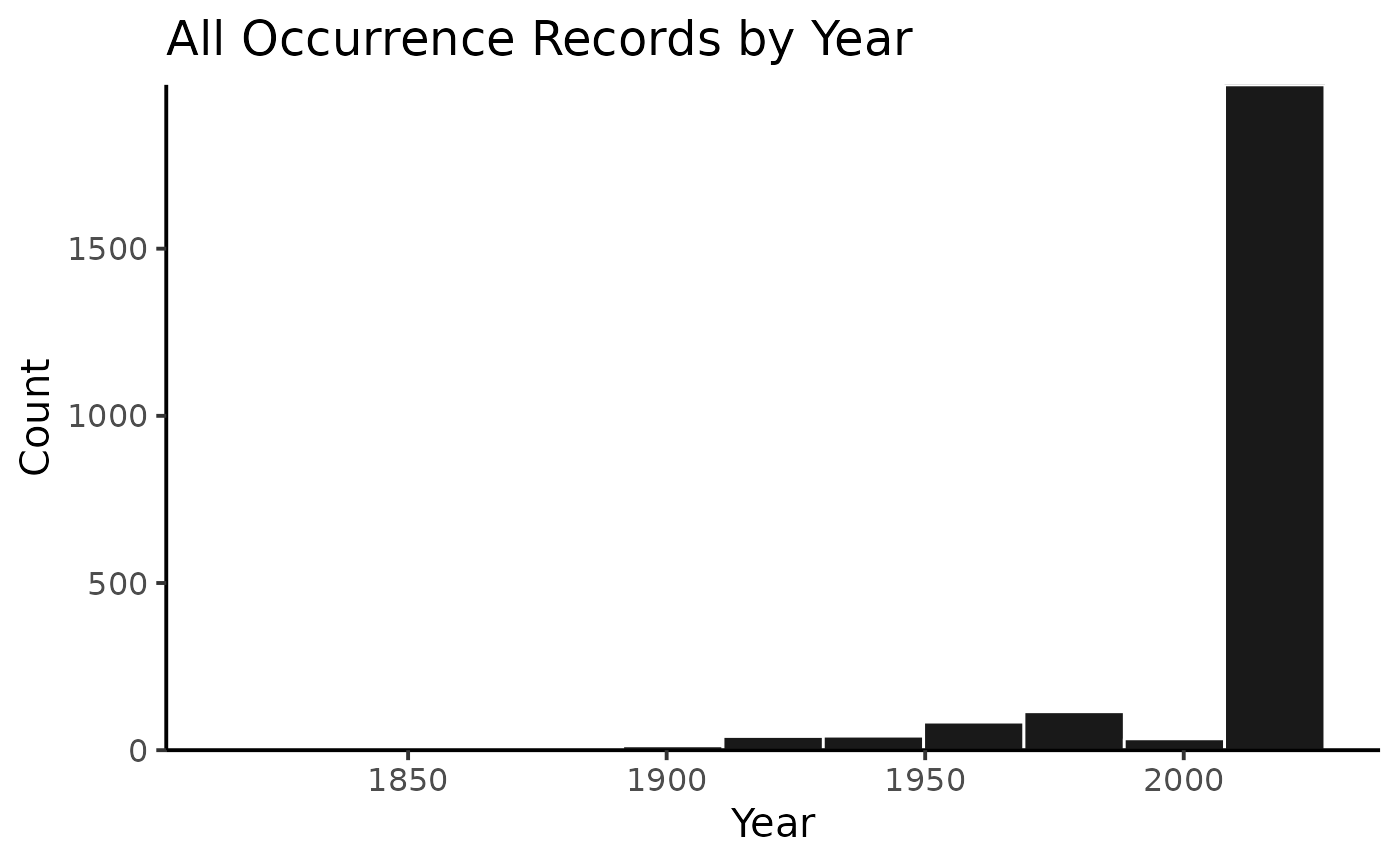
Simple citations
After doing a search for occurrence points, you can use
occCitation() to generate citations for primary
biodiversity databases, as well as database aggregators.
Note: Currently, GBIF and BIEN are the only aggregators
for which citations are supported.
#Get citations
mySimpleOccCitations <- occCitation(mySimpleOccCiteObject)Here is a simple way of generating a formatted citation document from
the results of occCitation().
print(mySimpleOccCitations)## Writing 5 Bibtex entries ... OK
## Results written to file 'temp.bib'## AFFOUARD A, JOLY A, LOMBARDO J, CHAMP J, GOEAU H, CHOUET M, GRESSE H, BONNET P (2025). Pl@ntNet observations. Version 1.9. Pl@ntNet. https://doi.org/10.15468/gtebaa. Accessed via GBIF on 2022-03-02.
## AFFOUARD A, JOLY A, LOMBARDO J, CHAMP J, GOEAU H, CHOUET M, GRESSE H, BOTELLA C, BONNET P (2023). Pl@ntNet automatically identified occurrences. Version 1.8. Pl@ntNet. https://doi.org/10.15468/mma2ec. Accessed via GBIF on 2022-03-02.
## Chamberlain, S., Barve, V., Mcglinn, D., Oldoni, D., Desmet, P., Geffert, L., Ram, K. (2026). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 3.8.4.5. https://CRAN.R-project.org/package = rgbif.
## Chamberlain, S., Boettiger, C. (2017). R Python, and Ruby clients for GBIF species occurrence data. PeerJ PrePrints.
## Fatima Parker-Allie, Ranwashe F (2018). PRECIS. South African National Biodiversity Institute. https://doi.org/10.15468/rckmn2. Accessed via GBIF on 2022-03-02.
## iNaturalist contributors, iNaturalist (2026). iNaturalist Research-grade Observations. iNaturalist.org. https://doi.org/10.15468/ab3s5x. Accessed via GBIF on 2022-03-02.
## Maitner, B. (2025). . R package version 1.2.7. NA.
## Missouri Botanical Garden,Herbarium. Accessed via BIEN on NA.
## MNHN, Chagnoux S (2025). The vascular plants collection (P) at the Herbarium of the Muséum national d'Histoire Naturelle (MNHN - Paris). Version 69.422. MNHN - Museum national d'Histoire naturelle. https://doi.org/10.15468/nc6rxy. Accessed via GBIF on 2022-03-02.
## MNHN. Accessed via BIEN on NA.
## naturgucker.de. NABU|naturgucker. https://doi.org/10.15468/uc1apo. Accessed via GBIF on 2022-03-02.
## Observation.org (2025). Observation.org, Nature data from around the World. https://doi.org/10.15468/5nilie. Accessed via GBIF on 2022-03-02.
## Owens, H., Merow, C., Maitner, B., Kass, J., Barve, V., Guralnick, R. (2026). occCite: Querying and Managing Large Biodiversity Occurrence Datasets. R package version 0.6.1. https://CRAN.R-project.org/package = occCite.
## Ranwashe F (2025). Botanical Database of Southern Africa (BODATSA): Botanical Collections. Version 1.30. South African National Biodiversity Institute. https://doi.org/10.15468/2aki0q. Accessed via GBIF on 2022-03-02.
## Rob Cubey (2022). Royal Botanic Garden Edinburgh Living Plant Collections (E). Royal Botanic Garden Edinburgh. https://doi.org/10.15468/bkzv1l. Accessed via GBIF on 2022-03-02.
## SANBI. Accessed via BIEN on NA.
## Senckenberg (2020). African Plants - a photo guide. https://doi.org/10.15468/r9azth. Accessed via GBIF on 2022-03-02.
## Taylor S (2019). G. S. Torrey Herbarium at the University of Connecticut (CONN). University of Connecticut. https://doi.org/10.15468/w35jmd. Accessed via GBIF on 2022-03-02.
## Team}, {.C. (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
## Teisher J, Stimmel H (2026). Tropicos MO Specimen Data. Missouri Botanical Garden. https://doi.org/10.15468/hja69f. Accessed via GBIF on 2022-03-02.
## Tela Botanica. Carnet en Ligne. https://doi.org/10.15468/rydcn2. Accessed via GBIF on 2022-03-02.
## UConn. Accessed via BIEN on NA.Simple Taxonomic Rectification
Note:The taxize package, which occCite
uses for taxonRectification(), has been archived. To
prevent occCite from being archived, which would result in
downstream problems, we have disabled external taxonomic rectification
as an option. If taxize comes back, or we identify an
alternative, we will reinstate this feature. The code still exists, it’s
just been commented out. Contact Hannah Owens (hannah.owens@gmail.com) for tips on how to reactivate
the feature using the gitHub version of taxize.
In the simplest of searches, such as the one above, the taxonomy of
your input species name is automatically rectified through the
occCite function studyTaxonList() using
gnr_resolve() from the taxize R
package. If you would like to change the source of the taxonomy being
used to rectify your species names, you can specify as many taxonomic
repositories as you like from the Global Names Index (GNI). The complete
list of GNI repositories can be found here.
studyTaxonList() chooses the taxonomic names closest to
those being input and documents which taxonomic repositories agreed with
those names. studyTaxonList() instantiates an
occCiteData object the same way occQuery()
does. This object can be passed into occQuery() to perform
your occurrence data search.
#Rectify taxonomy
myTROccCiteObject <- studyTaxonList(x = "Protea cynaroides",
datasources = c("National Center for Biotechnology Information",
"Encyclopedia of Life",
"Integrated Taxonomic Information SystemITIS"))
myTROccCiteObject@cleanedTaxonomyFor advanced features, please refer to
vignette("Advanced", package = "occCite").