
Advanced search and citation of occurrences
Hannah L. Owens
Cory Merow
Brian Maitner
Jamie M. Kass
Vijay Barve
Robert Guralnick
2026-02-15
Source:vignettes/b_Advanced.Rmd
b_Advanced.RmdAdvanced features
This vignette demonstrates more advanced features and customization
available in occCite. We recommend you read
vignette("Simple.Rmd", package = "occCite") first, if you
have not already done so.
Loading data from previous GBIF searches
Querying GBIF can take quite a bit of time, especially for multiple
species and/or well-known species. In this case, you may wish to access
previously-downloaded data sets from your computer by specifying the
general location of your downloaded .zip files.
occQuery will crawl through your specified
GBIFDownloadDirectory to collect all the .zip
files contained in that folder and its subfolders. It will then import
the most recent downloads that match your taxon list. These GBIF data
will be appended to a BIEN search the same as if you do the simple
real-time search (if you chose BIEN as well as GBIF), as was shown
above. checkPreviousGBIFDownload is TRUE by
default, but if loadLocalGBIFDownload is TRUE,
occQuery will ignore checkPreviousDownload. It
is also worth noting that occCite does not currently
support mixed data download sources. That is, you cannot do GBIF queries
for some taxa, download previously-prepared data sets for others, and
load the rest from local data sets on your computer.
# Simple search
myOldOccCiteObject <- occQuery(x = "Protea cynaroides",
datasources = c("gbif", "bien"),
GBIFLogin = GBIFLogin,
GBIFDownloadDirectory =
system.file('extdata/', package='occCite'),
checkPreviousGBIFDownload = T)Here is the result. Look familiar?
#GBIF search results
head(myOldOccCiteObject@occResults$`Protea cynaroides`$GBIF$OccurrenceTable);## name longitude latitude coordinateUncertaintyInMeters day month
## 1 Protea cynaroides 18.43928 -33.95440 8 17 2
## 2 Protea cynaroides 22.12754 -33.91561 4 11 2
## 3 Protea cynaroides 18.43927 -33.95429 8 17 2
## 4 Protea cynaroides 18.43254 -34.29275 31 6 2
## 5 Protea cynaroides 18.42429 -34.02934 2167 10 2
## 6 Protea cynaroides 18.43529 -34.10545 2 8 2
## year datasetKey dataService
## 1 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 2 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 3 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 4 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 5 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## 6 2022 50c9509d-22c7-4a22-a47d-8c48425ef4a7 GBIF
## datasetName
## 1 iNaturalist Research-grade Observations
## 2 iNaturalist Research-grade Observations
## 3 iNaturalist Research-grade Observations
## 4 iNaturalist Research-grade Observations
## 5 iNaturalist Research-grade Observations
## 6 iNaturalist Research-grade Observations
#The full summary
summary(myOldOccCiteObject)##
## OccCite query occurred on: 20 June, 2024
##
## User query type: User-supplied list of taxa.
##
## Sources for taxonomic rectification: GBIF Backbone Taxonomy
##
##
## Taxonomic cleaning results:
##
## Input Name Best Match Taxonomic Databases w/ Matches
## 1 Protea cynaroides Protea cynaroides (L.) L. GBIF Backbone Taxonomy
##
## Sources for occurrence data: gbif, bien
##
## Species Occurrences Sources
## 1 Protea cynaroides (L.) L. 2334 17
##
## GBIF dataset DOIs:
##
## Species GBIF Access Date GBIF DOI
## 1 Protea cynaroides (L.) L. 2022-03-02 10.15468/dl.ztbx8cGetting citation data works the exact same way with previously-downloaded data as it does from a fresh data set.
#Get citations
myOldOccCitations <- occCitation(myOldOccCiteObject)
print(myOldOccCitations)## Writing 5 Bibtex entries ... OK
## Results written to file 'temp.bib'## AFFOUARD A, JOLY A, LOMBARDO J, CHAMP J, GOEAU H, CHOUET M, GRESSE H, BONNET P (2025). Pl@ntNet observations. Version 1.9. Pl@ntNet. https://doi.org/10.15468/gtebaa. Accessed via GBIF on 2022-03-02.
## AFFOUARD A, JOLY A, LOMBARDO J, CHAMP J, GOEAU H, CHOUET M, GRESSE H, BOTELLA C, BONNET P (2023). Pl@ntNet automatically identified occurrences. Version 1.8. Pl@ntNet. https://doi.org/10.15468/mma2ec. Accessed via GBIF on 2022-03-02.
## Chamberlain, S., Barve, V., Mcglinn, D., Oldoni, D., Desmet, P., Geffert, L., Ram, K. (2026). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 3.8.4.5. https://CRAN.R-project.org/package = rgbif.
## Chamberlain, S., Boettiger, C. (2017). R Python, and Ruby clients for GBIF species occurrence data. PeerJ PrePrints.
## Fatima Parker-Allie, Ranwashe F (2018). PRECIS. South African National Biodiversity Institute. https://doi.org/10.15468/rckmn2. Accessed via GBIF on 2022-03-02.
## iNaturalist contributors, iNaturalist (2026). iNaturalist Research-grade Observations. iNaturalist.org. https://doi.org/10.15468/ab3s5x. Accessed via GBIF on 2022-03-02.
## Maitner, B. (2025). . R package version 1.2.7. NA.
## Missouri Botanical Garden,Herbarium. Accessed via BIEN on NA.
## MNHN, Chagnoux S (2025). The vascular plants collection (P) at the Herbarium of the Muséum national d'Histoire Naturelle (MNHN - Paris). Version 69.422. MNHN - Museum national d'Histoire naturelle. https://doi.org/10.15468/nc6rxy. Accessed via GBIF on 2022-03-02.
## MNHN. Accessed via BIEN on NA.
## naturgucker.de. NABU|naturgucker. https://doi.org/10.15468/uc1apo. Accessed via GBIF on 2022-03-02.
## Observation.org (2025). Observation.org, Nature data from around the World. https://doi.org/10.15468/5nilie. Accessed via GBIF on 2022-03-02.
## Owens, H., Merow, C., Maitner, B., Kass, J., Barve, V., Guralnick, R. (2026). occCite: Querying and Managing Large Biodiversity Occurrence Datasets. R package version 0.6.1. https://CRAN.R-project.org/package = occCite.
## Ranwashe F (2025). Botanical Database of Southern Africa (BODATSA): Botanical Collections. Version 1.30. South African National Biodiversity Institute. https://doi.org/10.15468/2aki0q. Accessed via GBIF on 2022-03-02.
## Rob Cubey (2022). Royal Botanic Garden Edinburgh Living Plant Collections (E). Royal Botanic Garden Edinburgh. https://doi.org/10.15468/bkzv1l. Accessed via GBIF on 2022-03-02.
## SANBI. Accessed via BIEN on NA.
## Senckenberg (2020). African Plants - a photo guide. https://doi.org/10.15468/r9azth. Accessed via GBIF on 2022-03-02.
## Taylor S (2019). G. S. Torrey Herbarium at the University of Connecticut (CONN). University of Connecticut. https://doi.org/10.15468/w35jmd. Accessed via GBIF on 2022-03-02.
## Team}, {.C. (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
## Teisher J, Stimmel H (2026). Tropicos MO Specimen Data. Missouri Botanical Garden. https://doi.org/10.15468/hja69f. Accessed via GBIF on 2022-03-02.
## Tela Botanica. Carnet en Ligne. https://doi.org/10.15468/rydcn2. Accessed via GBIF on 2022-03-02.
## UConn. Accessed via BIEN on NA.Note that you can also load multiple species using either a vector of species names or a phylogeny (provided you have previously downloaded data for all of the species of interest), and you can load occurrences from non-GBIF data sources (e.g. BIEN) in the same query.
Performing a Multi-Species Search
In addition to doing a simple, single species search, you can also
use occCite to search for and manage occurrence datasets
for multiple species. You can either submit a vector of species names,
or you can submit a phylogeny! The occCitation function will
return a named list of citation tables in the case of multiple
species.
occCite with a Phylogeny
Here is an example of how such a search is structured, using an unpublished phylogeny of billfishes.
library(ape)
#Get tree
treeFile <- system.file("extdata/Fish_12Tax_time_calibrated.tre", package='occCite')
phylogeny <- ape::read.nexus(treeFile)
tree <- ape::extract.clade(phylogeny, 22)
#Query databases for names
myPhyOccCiteObject <- studyTaxonList(x = tree,
datasources = "GBIF Backbone Taxonomy")## handled warning: Following sources not found in
## Global Names Index source list: GBIF Backbone Taxonomy## handled warning: No valid taxonomic data sources supplied.
## Populating default list from all available sources.## handled warning: Following sources not found in
## Global Names Index source list: GBIF Backbone Taxonomy## handled warning: No valid taxonomic data sources supplied.
## Populating default list from all available sources.## handled warning: Following sources not found in
## Global Names Index source list: GBIF Backbone Taxonomy## handled warning: No valid taxonomic data sources supplied.
## Populating default list from all available sources.
#Query GBIF for occurrence data
myPhyOccCiteObject <- occQuery(x = myPhyOccCiteObject,
datasources = "gbif",
GBIFDownloadDirectory = system.file('extdata/', package='occCite'),
loadLocalGBIFDownload = T,
checkPreviousGBIFDownload = F)
# What does a multispecies query look like?
summary(myPhyOccCiteObject)##
## OccCite query occurred on: 15 February, 2026
##
## User query type: User-supplied phylogeny.
##
## Sources for taxonomic rectification: GBIF Backbone Taxonomy
##
##
## Taxonomic cleaning results:
##
## Input Name Best Match
## 1 Tetrapturus_angustirostris Tetrapturus angustirostris Tanaka, 1915
## 2 Tetrapturus_belone Tetrapturus belone Rafinesque, 1810
## 3 Tetrapturus_pfluegeri Tetrapturus pfluegeri Robins & de Sylva, 1963
## Taxonomic Databases w/ Matches
## 1 Catalogue of Life
## 2 Catalogue of Life
## 3 Catalogue of Life
##
## Sources for occurrence data: gbif
##
## Species Occurrences Sources
## 1 Tetrapturus angustirostris Tanaka, 1915 649 23
## 2 Tetrapturus belone Rafinesque, 1810 9 6
## 3 Tetrapturus pfluegeri Robins & de Sylva, 1963 410 8
##
## GBIF dataset DOIs:
##
## Species GBIF Access Date
## 1 Tetrapturus angustirostris Tanaka, 1915 2019-07-04
## 2 Tetrapturus belone Rafinesque, 1810 2019-07-04
## 3 Tetrapturus pfluegeri Robins & de Sylva, 1963 2019-07-04
## GBIF DOI
## 1 10.15468/dl.mumi5e
## 2 10.15468/dl.q2nxb1
## 3 10.15468/dl.qjidbsWhen you have results for multiple species, as in this case, you can also plot the summary figures either for the whole search…
plot(myPhyOccCiteObject)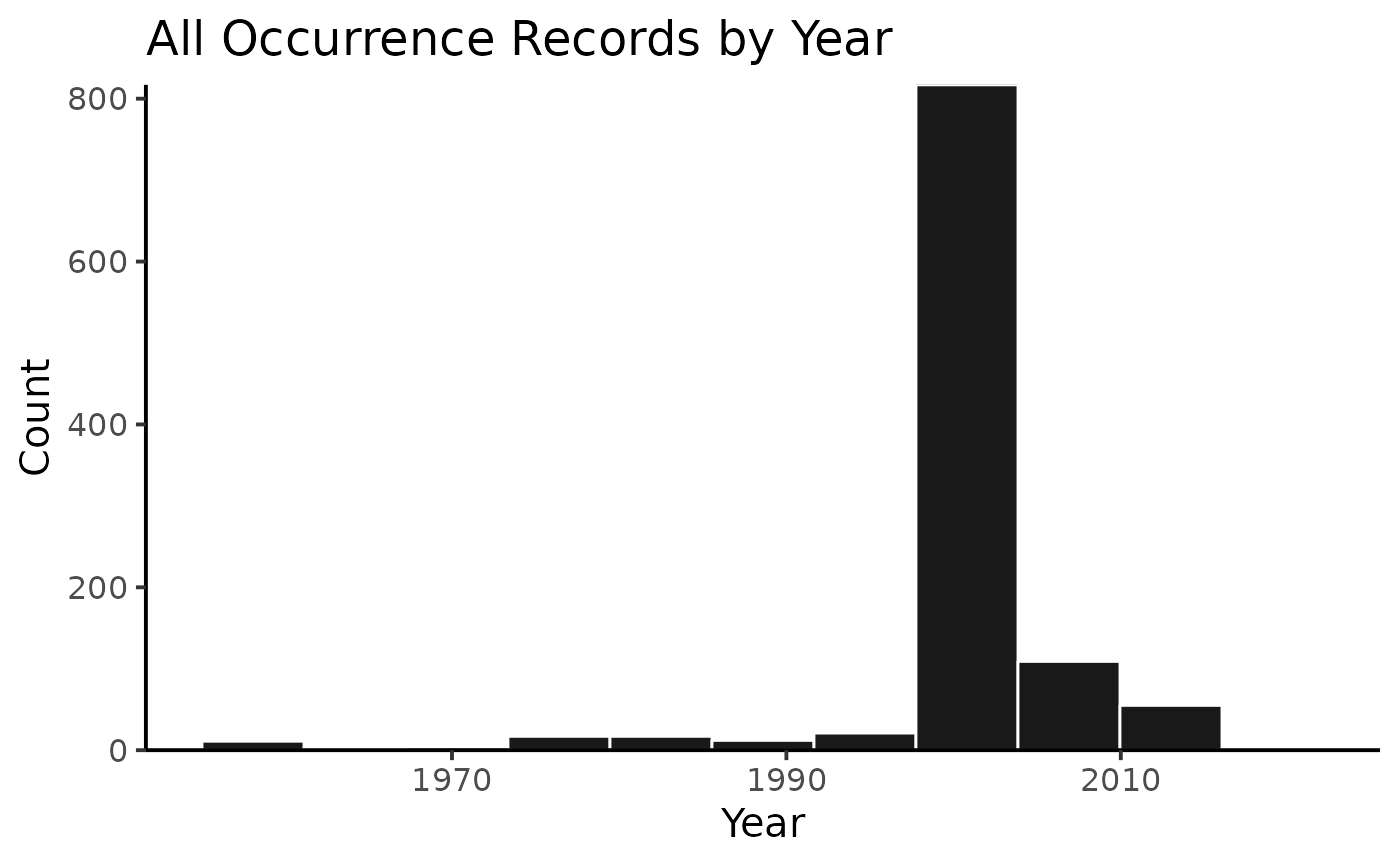
or you can plot the results by species!
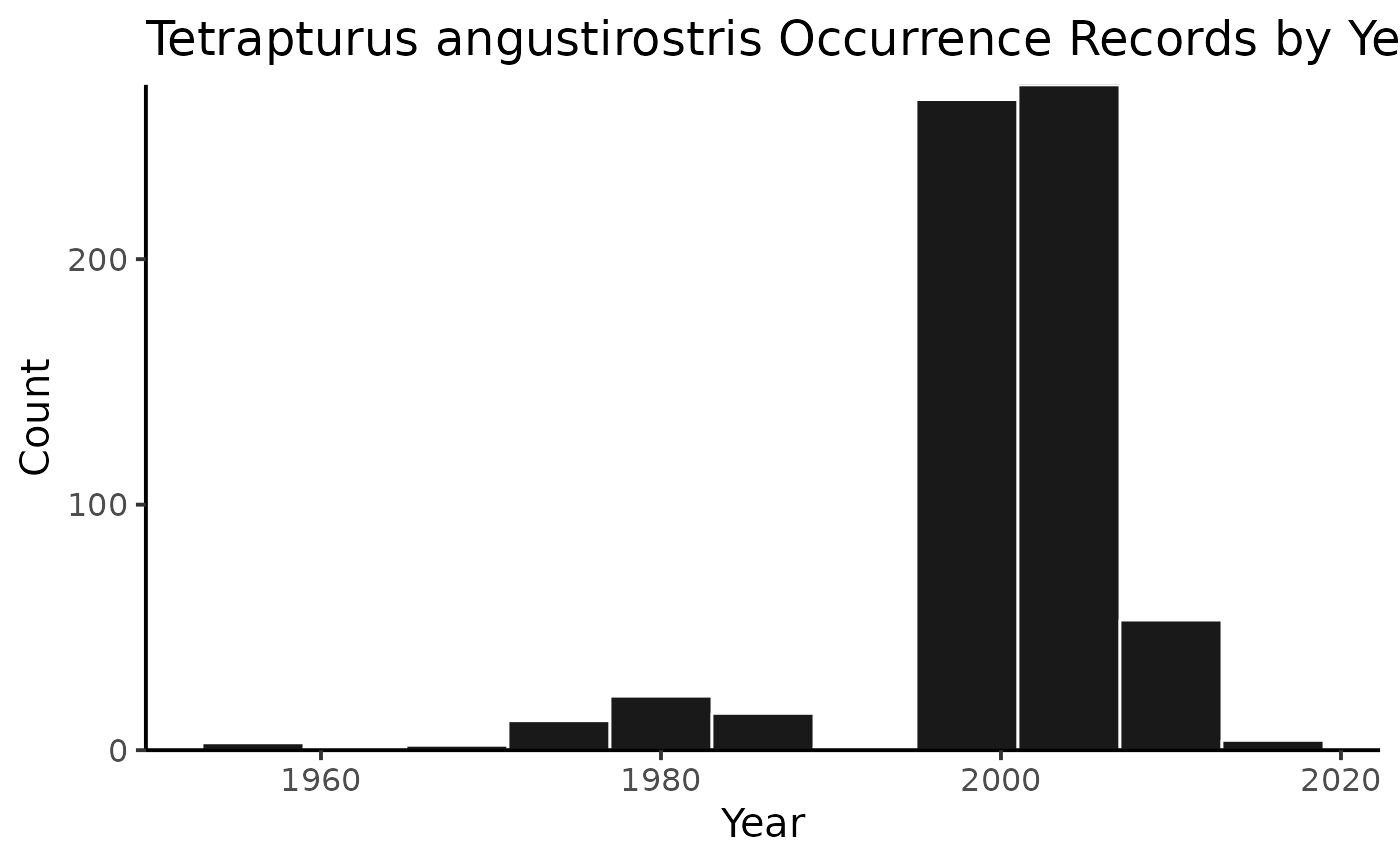
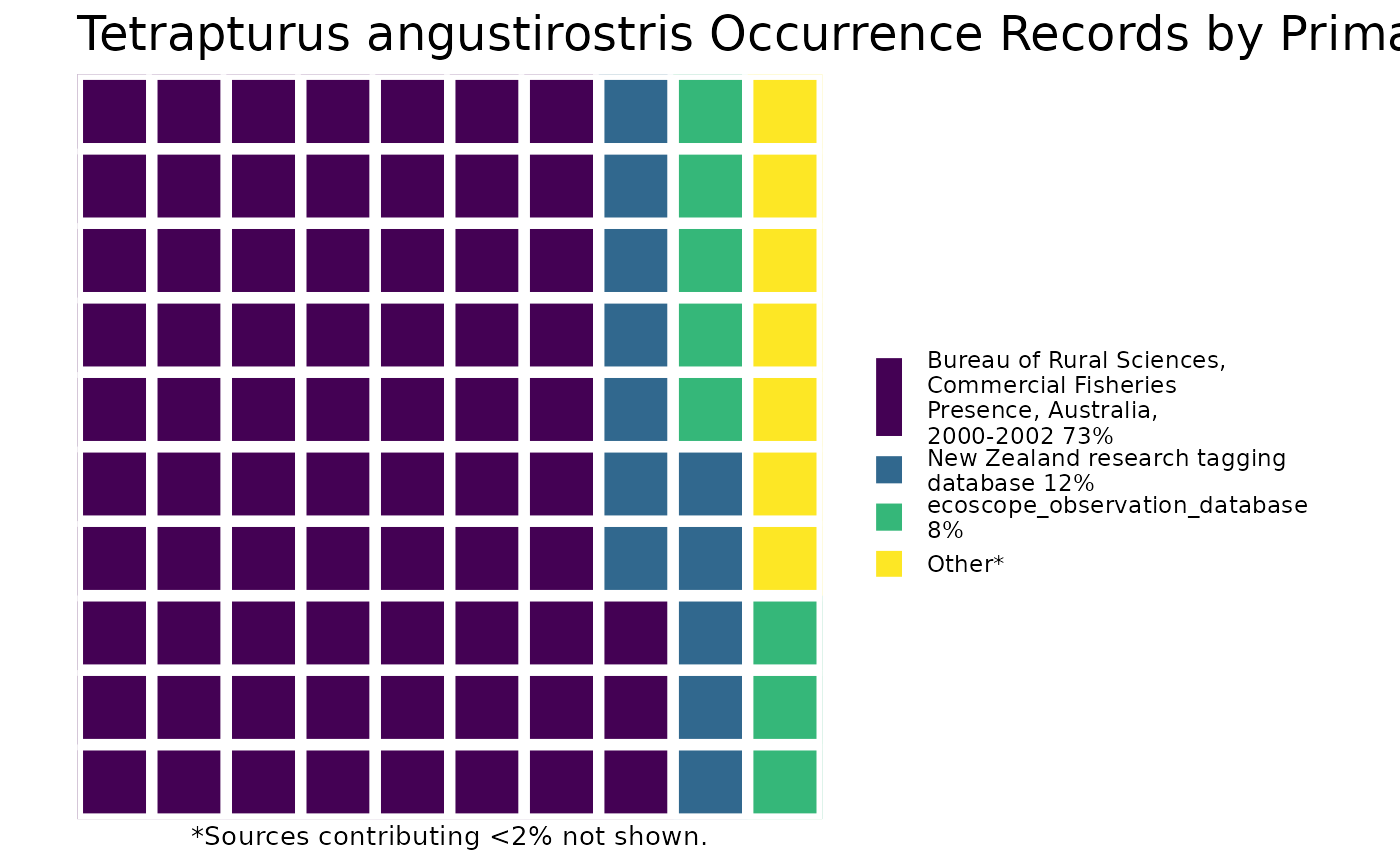
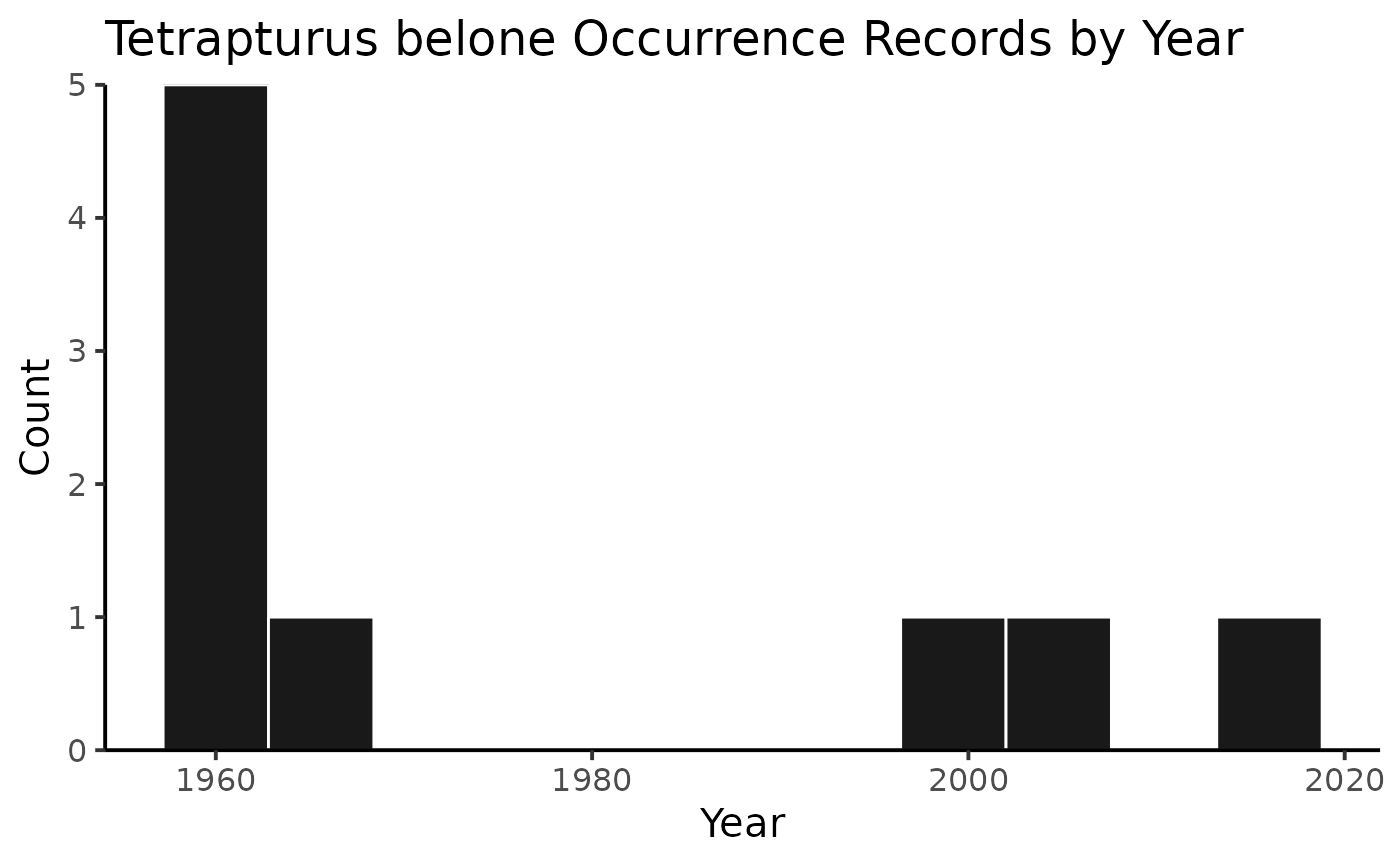
And then you can print out the citations, separated by species (or not, but in this example, they’re separate).
#Get citations
myPhyOccCitations <- occCitation(myPhyOccCiteObject)
#Print citations as text with accession dates.
print(myPhyOccCitations, bySpecies = T)## Writing 6 Bibtex entries ... OK
## Results written to file 'temp.bib'## Species: Tetrapturus angustirostris Tanaka, 1915
##
## Australian Museum (2026). Australian Museum provider for OZCAM. https://doi.org/10.15468/e7susi. Accessed via GBIF on 2019-07-04.
## Barde J (2011). ecoscope_observation_database. IRD - Institute of Research for Development. https://doi.org/10.15468/dz1kk0. Accessed via GBIF on 2019-07-04.
## Bureau of Rural Sciences - National commercial fisheries half-degree data set 2000-2002 https://doi.org/10.15468/0esdv0. Accessed via GBIF on 2019-07-04.
## Cauquil P, Barde J (2011). observe_tuna_bycatch_ecoscope. IRD - Institute of Research for Development. https://doi.org/10.15468/23m361. Accessed via GBIF on 2019-07-04.
## Chamberlain, S., Barve, V., Mcglinn, D., Oldoni, D., Desmet, P., Geffert, L., Ram, K. (2026). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 3.8.4.5. https://CRAN.R-project.org/package = rgbif.
## Chamberlain, S., Boettiger, C. (2017). R Python, and Ruby clients for GBIF species occurrence data. PeerJ PrePrints.
## Chiang W (2014). Taiwan Fisheries Research Institute – Digital archives of coastal and offshore specimens. TELDAP. https://doi.org/10.15468/xvxngy. Accessed via GBIF on 2019-07-04.
## European Nucleotide Archive (EMBL-EBI) (2019). Geographically tagged INSDC sequences. https://doi.org/10.15468/cndomv. Accessed via GBIF on 2019-07-04.
## Fong J (2026). CAS Ichthyology (ICH). Version 150.513. California Academy of Sciences. Occurrence dataset. http://ipt.calacademy.org:8080/resource?r=ich&v=150.513 https://doi.org/10.15468/efh2ib. Accessed via GBIF on 2019-07-04.
## Frable B (2025). SIO Marine Vertebrate Collection. Version 1.9. Scripps Institution of Oceanography. https://doi.org/10.15468/ad1ovc. Accessed via GBIF on 2019-07-04.
## Harvard University M, Morris P J (2026). Museum of Comparative Zoology, Harvard University. Version 162.500. Museum of Comparative Zoology, Harvard University. https://doi.org/10.15468/p5rupv. Accessed via GBIF on 2019-07-04.
## iNaturalist contributors, iNaturalist (2026). iNaturalist Research-grade Observations. iNaturalist.org. https://doi.org/10.15468/ab3s5x. Accessed via GBIF on 2019-07-04.
## Inventaire National du Patrimoine Naturel (2018). Programme Ecoscope: données d'observations des écosystèmes marins exploités (Réunion). UAR PatriNat (OFB-MNHN-CNRS-IRD), Paris. https://doi.org/10.15468/elttrd. Accessed via GBIF on 2019-07-04.
## Inventaire National du Patrimoine Naturel (2018). Programme Ecoscope: données d'observations des écosystèmes marins exploités. UAR PatriNat (OFB-MNHN-CNRS-IRD), Paris. https://doi.org/10.15468/gdrknh. Accessed via GBIF on 2019-07-04.
## McLean, M.W. (2014). Straightforward Bibliography Management in R Using the RefManager Package. NA, NA. https://arxiv.org/abs/1403.2036.
## McLean, M.W. (2017). RefManageR: Import and Manage BibTeX and BibLaTeX References in R. The Journal of Open Source Software.
## Mertz W, Ludt W, Clardy T, Robson S, Camacho N (2025). LACM Vertebrate Collection. Version 18.19. Natural History Museum of Los Angeles County. https://doi.org/10.15468/77rmwd. Accessed via GBIF on 2019-07-04.
## Ministry for Primary Industries (2014). New Zealand research tagging database. Southwestern Pacific OBIS, National Institute of Water and Atmospheric Research (NIWA), Wellington, New Zealand, 411926 records, Online http://nzobisipt.niwa.co.nz/resource.do?r=mpi_tag released on November 5, 2014. https://doi.org/10.15468/i66xdm. Accessed via GBIF on 2019-07-04.
## Motomura H (2025). Fish collection of the Kagoshima University Museum. National Museum of Nature and Science, Japan. https://doi.org/10.15468/vcj3j8. Accessed via GBIF on 2019-07-04.
## Owens, H., Merow, C., Maitner, B., Kass, J., Barve, V., Guralnick, R. (2026). occCite: Querying and Managing Large Biodiversity Occurrence Datasets. R package version 0.6.1. https://CRAN.R-project.org/package = occCite.
## Queensland Museum (2026). Queensland Museum provider for OZCAM. https://doi.org/10.15468/lotsye. Accessed via GBIF on 2019-07-04.
## Raiva R, Santana P (2021). Diversidade e ocorrência de peixes em Inhambane (2009-2017). Version 1.7. National Institute of Fisheries Research (IIP) – Mozambique. https://doi.org/10.15468/4fj2tq. Accessed via GBIF on 2019-07-04.
## Raiva R, Viador R, Santana P (2021). Diversidade e ocorrência de peixes na Zambézia (2003-2016). Version 1.6. National Institute of Fisheries Research (IIP) – Mozambique. https://doi.org/10.15468/mrz36h. Accessed via GBIF on 2019-07-04.
## Robins R (2026). UF FLMNH Ichthyology. Version 117.536. Florida Museum of Natural History. https://doi.org/10.15468/8mjsel. Accessed via GBIF on 2019-07-04.
## South Australian Museum (2025). South Australian Museum Adelaide provider for OZCAM. https://doi.org/10.15468/wz4rrh. Accessed via GBIF on 2019-07-04.
## Team}, {.C. (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
## The International Barcode of Life Consortium (2026). International Barcode of Life project (iBOL). https://doi.org/10.15468/inygc6. Accessed via GBIF on 2019-07-04.
## Uchifune Y, Yamamoto H (2024). Asia-Pacific Dataset. Version 1.37. National Museum of Nature and Science, Japan. https://doi.org/10.48518/00002. Accessed via GBIF on 2019-07-04.
## Western Australian Museum (2024). Western Australian Museum provider for OZCAM. https://doi.org/10.15468/5qt0dm. Accessed via GBIF on 2019-07-04.## Writing 6 Bibtex entries ... OK
## Results written to file 'temp.bib'## Species: Tetrapturus belone Rafinesque, 1810
##
## Chamberlain, S., Barve, V., Mcglinn, D., Oldoni, D., Desmet, P., Geffert, L., Ram, K. (2026). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 3.8.4.5. https://CRAN.R-project.org/package = rgbif.
## Chamberlain, S., Boettiger, C. (2017). R Python, and Ruby clients for GBIF species occurrence data. PeerJ PrePrints.
## Conselleria de Medio Ambiente, Agua, Infraestructuras y Territorio. Generalitat Valenciana (2024). Banco de Datos de la Biodiversidad de la Comunitat Valenciana. Biodiversity data bank of Generalitat Valenciana. https://doi.org/10.15468/b4yqdy. Accessed via GBIF on 2019-07-04.
## European Nucleotide Archive (EMBL-EBI) (2019). Geographically tagged INSDC sequences. https://doi.org/10.15468/cndomv. Accessed via GBIF on 2019-07-04.
## Harvard University M, Morris P J (2026). Museum of Comparative Zoology, Harvard University. Version 162.500. Museum of Comparative Zoology, Harvard University. https://doi.org/10.15468/p5rupv. Accessed via GBIF on 2019-07-04.
## McLean, M.W. (2014). Straightforward Bibliography Management in R Using the RefManager Package. NA, NA. https://arxiv.org/abs/1403.2036.
## McLean, M.W. (2017). RefManageR: Import and Manage BibTeX and BibLaTeX References in R. The Journal of Open Source Software.
## Owens, H., Merow, C., Maitner, B., Kass, J., Barve, V., Guralnick, R. (2026). occCite: Querying and Managing Large Biodiversity Occurrence Datasets. R package version 0.6.1. https://CRAN.R-project.org/package = occCite.
## ROBERT S, LEPAREUR F, Inventaire National du Patrimoine Naturel (2022). Données d'occurrences Espèces issues de l'inventaire des ZNIEFF. Version 1.7. UAR PatriNat (OFB-MNHN-CNRS-IRD), Paris. https://doi.org/10.15468/ikshke. Accessed via GBIF on 2019-07-04.
## Robins R (2026). UF FLMNH Ichthyology. Version 117.536. Florida Museum of Natural History. https://doi.org/10.15468/8mjsel. Accessed via GBIF on 2019-07-04.
## Team}, {.C. (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
## University of Kansas Biodiversity Institute: KUBI Ichthyology Collection https://doi.org/10.15468/mgjasg. Accessed via GBIF on 2019-07-04.## Writing 6 Bibtex entries ... OK
## Results written to file 'temp.bib'## Species: Tetrapturus pfluegeri Robins & de Sylva, 1963
##
## Barde J (2011). ecoscope_observation_database. IRD - Institute of Research for Development. https://doi.org/10.15468/dz1kk0. Accessed via GBIF on 2019-07-04.
## Boateng M (2021). Fishes of Ghana. Version 1.4. Department of Marine and Fisheries Sciences, University of Ghana. https://doi.org/10.15468/pgesnw. Accessed via GBIF on 2019-07-04.
## Cauquil P, Barde J (2011). observe_tuna_bycatch_ecoscope. IRD - Institute of Research for Development. https://doi.org/10.15468/23m361. Accessed via GBIF on 2019-07-04.
## Chamberlain, S., Barve, V., Mcglinn, D., Oldoni, D., Desmet, P., Geffert, L., Ram, K. (2026). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 3.8.4.5. https://CRAN.R-project.org/package = rgbif.
## Chamberlain, S., Boettiger, C. (2017). R Python, and Ruby clients for GBIF species occurrence data. PeerJ PrePrints.
## European Nucleotide Archive (EMBL-EBI) (2019). Geographically tagged INSDC sequences. https://doi.org/10.15468/cndomv. Accessed via GBIF on 2019-07-04.
## Inventaire National du Patrimoine Naturel (2018). Programme Ecoscope: données d'observations des écosystèmes marins exploités (Réunion). UAR PatriNat (OFB-MNHN-CNRS-IRD), Paris. https://doi.org/10.15468/elttrd. Accessed via GBIF on 2019-07-04.
## Inventaire National du Patrimoine Naturel (2018). Programme Ecoscope: données d'observations des écosystèmes marins exploités. UAR PatriNat (OFB-MNHN-CNRS-IRD), Paris. https://doi.org/10.15468/gdrknh. Accessed via GBIF on 2019-07-04.
## McLean, M.W. (2014). Straightforward Bibliography Management in R Using the RefManager Package. NA, NA. https://arxiv.org/abs/1403.2036.
## McLean, M.W. (2017). RefManageR: Import and Manage BibTeX and BibLaTeX References in R. The Journal of Open Source Software.
## Owens, H., Merow, C., Maitner, B., Kass, J., Barve, V., Guralnick, R. (2026). occCite: Querying and Managing Large Biodiversity Occurrence Datasets. R package version 0.6.1. https://CRAN.R-project.org/package = occCite.
## Robins R (2026). UF FLMNH Ichthyology. Version 117.536. Florida Museum of Natural History. https://doi.org/10.15468/8mjsel. Accessed via GBIF on 2019-07-04.
## Team}, {.C. (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
## The International Barcode of Life Consortium (2026). International Barcode of Life project (iBOL). https://doi.org/10.15468/inygc6. Accessed via GBIF on 2019-07-04.