Following the template in OpenAlex’s oa-percentage tutorial, this vignette uses openalexR to answer:
How many of recent journal articles from the University of Pennsylvania are open access? And how many aren’t?
We first need to find the openalex.id
for University of Pennsylvania. We can do this by fetching for the
institutions entity and put “University of
Pennsylvania” in display_name or
display_name.search:
oa_fetch(
entity = "inst", # same as "institutions"
display_name.search = "\"University of Pennsylvania\""
) %>%
select(display_name, ror) %>%
knitr::kable()| display_name | ror |
|---|---|
| University of Pennsylvania | https://ror.org/00b30xv10 |
| California University of Pennsylvania | https://ror.org/01spssf70 |
| Hospital of the University of Pennsylvania | https://ror.org/02917wp91 |
| University of Pennsylvania Health System | https://ror.org/04h81rw26 |
| Indiana University of Pennsylvania | https://ror.org/0511cmw96 |
| University of Pennsylvania Press | https://ror.org/03xwa9562 |
| Cheyney University of Pennsylvania | https://ror.org/02nckwn80 |
We will use the first ror, 00b30xv10, as one of the filters for our query.
Alternatively, we could go to the autocomplete endpoint at https://explore.openalex.org/ to search for “University of Pennsylvania” and find the ror there!
All other filters are straightforward and explained in detailed in
the original jupyter notebook tutorial.
The only difference here is that, instead of grouping by
is_oa, we’re interested in the “trend” over the years, so
we’re going to group by publication_year, and perform the
query twice, one for is_oa = "true" and one for
is_oa = "false" .
open_access <- oa_fetch(
entity = "works",
institutions.ror = "00b30xv10",
type = "article",
from_publication_date = "2012-08-24",
is_paratext = "false",
is_oa = "true",
group_by = "publication_year"
)
closed_access <- oa_fetch(
entity = "works",
institutions.ror = "00b30xv10",
type = "article",
from_publication_date = "2012-08-24",
is_paratext = "false",
is_oa = "false",
group_by = "publication_year"
)
uf_df <- closed_access %>%
select(- key_display_name) %>%
full_join(open_access, by = "key", suffix = c("_ca", "_oa"))
uf_df
#> key count_ca key_display_name count_oa
#> 1 2018 4300 2018 5447
#> 2 2025 4131 2025 6956
#> 3 2022 4127 2022 7327
#> 4 2019 4091 2019 6220
#> 5 2020 4081 2020 7705
#> 6 2021 4030 2021 7842
#> 7 2024 3873 2024 7705
#> 8 2015 3829 2015 4528
#> 9 2014 3779 2014 4405
#> 10 2013 3677 2013 4380
#> 11 2016 3636 2016 4845
#> 12 2017 3592 2017 5177
#> 13 2023 3489 2023 8189
#> 14 2012 1156 2012 1148
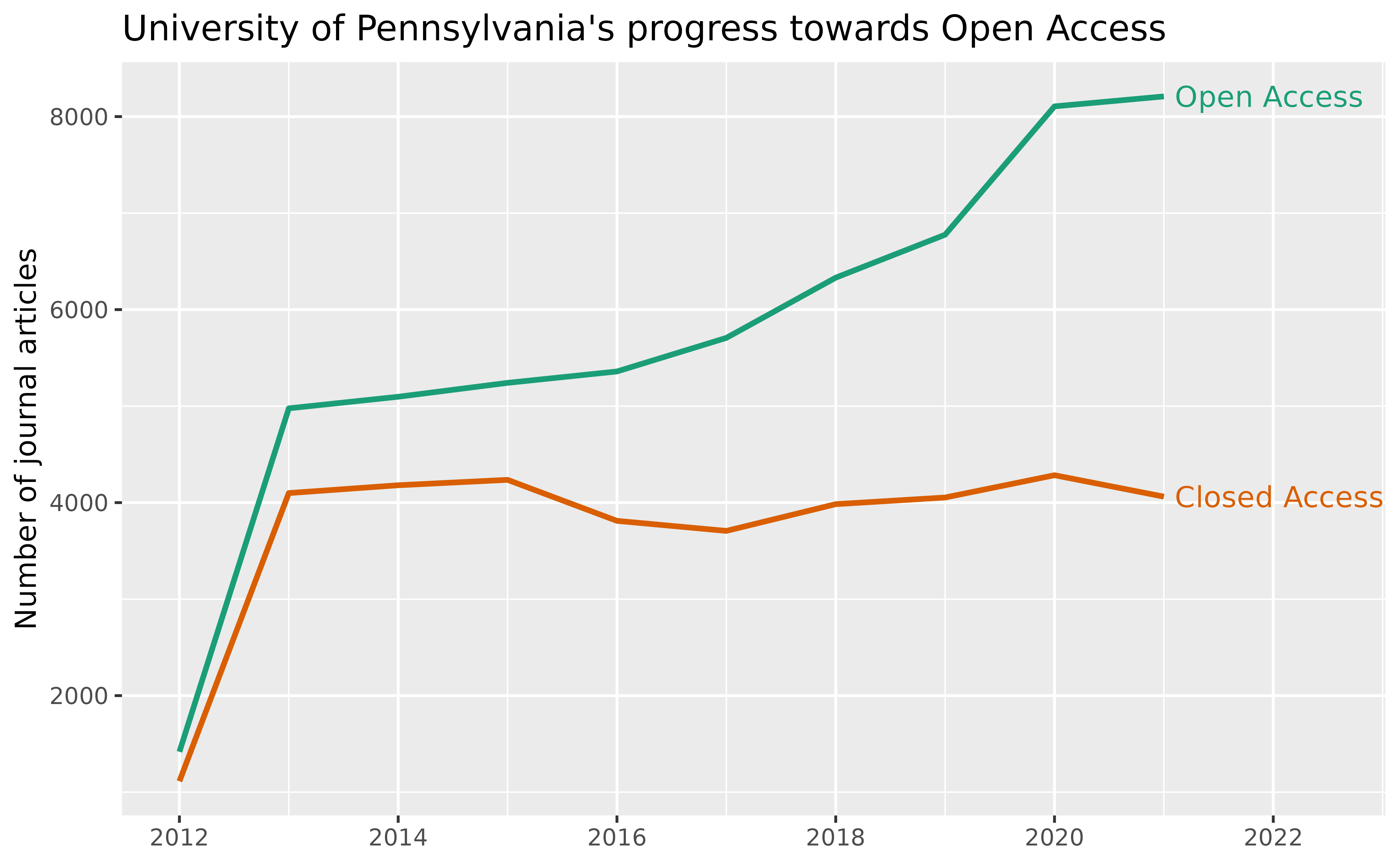
#> 15 2026 305 2026 487Finally, we compare the number of open vs. closed access articles over the years:
uf_df %>%
filter(key <= 2021) %>% # we do not yet have complete data for 2022 and after
pivot_longer(cols = starts_with("count")) %>%
mutate(
year = as.integer(key),
is_oa = recode(
name,
"count_ca" = "Closed Access",
"count_oa" = "Open Access"
),
label = if_else(key < 2021, NA_character_, is_oa)
) %>%
select(year, value, is_oa, label) %>%
ggplot(aes(x = year, y = value, group = is_oa, color = is_oa)) +
geom_line(size = 1) +
labs(
title = "University of Pennsylvania's progress towards Open Access",
x = NULL, y = "Number of journal articles") +
scale_color_brewer(palette = "Dark2", direction = -1) +
scale_x_continuous(breaks = seq(2010, 2024, 2)) +
geom_text(aes(label = label), nudge_x = 0.1, hjust = 0) +
coord_cartesian(xlim = c(NA, 2022.5)) +
guides(color = "none")