Performance and optimization
Source:vignettes/articles/performance-optimization.Rmd
performance-optimization.RmdWhile oa_fetch() offers a convenient and flexible way of
retrieving results from queries to the OpenAlex API, its defaults may
not be best suited for heavier workflows that involve fetching records
in the magnitude of tens or hundreds of thousands of entities.
Optimizing the performance of such large queries benefits greatly from being intentional and specific about what kinds of information you care about, and making assumptions that let you safely take shortcuts around the defaults.
This vignette discusses three strategies for for optimizing performance of large queries:
- The parameter
options = list(select = ...)inoa_fetch() - The argument
output = "list"inoa_fetch() - The function
oa_generate()
The select strategy
The options argument of oa_fetch()
specifies a list of additional parameters to add to the query, such as
select, sort, sample, and
seed. Of these, select can be used to specify
which fields of the entities are to be returned by OpenAlex. By
specifying only the kinds of information about entities that you care
about, you can reduce the overall size of the query result, which will
in turn speed up the fetching of the raw JSON and its conversion to a
data frame.
For example, suppose that we are looking for a sample of works from
the Topic of
Language Development and
Acquisition in Children ("T10730").
language_development <- oa_fetch(
entity = "topics",
search = "Language Development and Acquisition in Children"
)[1,1:2]
language_development
#> # A tibble: 1 × 2
#> id display_name
#> <chr> <chr>
#> 1 https://openalex.org/T10730 Language Development and DisordersTo sample some papers from this topic, we can use the
topics.id filter
and set options = list(sample = 5, seed = 1) to return a
random set of five Works entities
with a reproducible seed:
oa_fetch(
entity = "works",
topics.id = language_development$id,
options = list(sample = 5, seed = 1)
) %>%
show_works()
#> # A tibble: 5 × 6
#> id display_name first_author last_author is_oa top_concepts
#> <chr> <chr> <chr> <chr> <lgl> <chr>
#> 1 W2969728018 Examination of Metali… Amanda Krie… NA TRUE Metalinguis…
#> 2 W2371085312 Trastornos cognitivos… Valeria Abu… Aldo Ferre… FALSE Human immun…
#> 3 W2962551760 Conversations with a … Colwyn Trev… NA FALSE Action (phy…
#> 4 W2055502576 Preserved category le… Catherine H… Michael Co… FALSE Amnesia, Re…
#> 5 W2045105551 The Use of Profanity … John M. Rin… Po H. Lu FALSE Fluency, Ap…In OpenAlex, entities have a set of fields which
represent various information about them. These are typically returned
as data frame columns by oa_fetch(), and the full list of
fields can be found in the API documentation for each entity. For
example, the
fields in a Works object contain information such as
id, display_name, authorships,
and so on.
If we only cared about the above three fields from our sample of
papers, we can simplify specify those fields in the select
parameters of the options list of arguments:
oa_fetch(
entity = "works",
topics.id = language_development$id,
options = list(sample = 5, seed = 1,
select = c("id", "display_name", "authorships"))
)
#> # A tibble: 5 × 3
#> id display_name authorships
#> <chr> <chr> <list>
#> 1 https://openalex.org/W2969728018 Examination of Metalinguistic Sk… <tibble>
#> 2 https://openalex.org/W2371085312 Trastornos cognitivos en pacient… <tibble>
#> 3 https://openalex.org/W2962551760 Conversations with a two-month-o… <tibble>
#> 4 https://openalex.org/W2055502576 Preserved category learning in A… <tibble>
#> 5 https://openalex.org/W2045105551 The Use of Profanity During Lett… <tibble>This returns the scalar fields id and
display_name in their appropriate data types (character) in
the dataframe. Additionally, the authorships field has been
further processed as a list-column of data frames, to fit nicely into
the “tidy” data frame structure.
Specifying the desired fields up front in this way is not only
convenient but also more performant, as there will be less data for
oa_fetch() to process.
The output = "list" strategy
By default, oa_fetch() uses
output = "tibble", which returns a processed
tibble data frame of the results. In such cases, the JSON
response from OpenAlex is first converted to an R list, then a data
frame via oa2df(), which calls the appropriate conversion
implementation depending on the type of entity being processed (e.g.,
works2df() for Works entities).
A lot of care goes into oa2df() to return a compact,
tidy-data representation of query results. But these operations can
become a bottleneck to performance at scale, and so sometimes you may
want to opt out of this automatic data frame conversion.
To do so in oa_fetch(), you can set
output = "list", which will simply return the R list
corresponding to the JSON response.
output_list <- oa_fetch(
entity = "works",
topics.id = language_development$id,
options = list(sample = 5, seed = 1),
output = "list"
)
str(output_list, max.level = 1)
#> List of 5
#> $ :List of 50
#> $ :List of 50
#> $ :List of 50
#> $ :List of 50
#> $ :List of 50The list output can get quite unruly — each record contains dozens of
fields, some of which may be multiply nested. Moreover, some records may
have missing or incomplete fields, so extra care must be taken with the
output = "list" approach.
One advantage of returning the output as a list is that you can
always come back to process them as data frames later. Instead of
retrieving and converting the results simultaneously, which may
stress oa_fetch() for large queries, you can retrieve all
the results first and then convert them after the fact.
In our case, the Works entities can be processed with
works2df() (or more generally,
oa2df(entity = "works")), which returns a data frame
identical to what we saw at the start with the default
output = "tibble":
works2df(output_list) %>%
show_works()
#> # A tibble: 5 × 6
#> id display_name first_author last_author is_oa top_concepts
#> <chr> <chr> <chr> <chr> <lgl> <chr>
#> 1 W2969728018 Examination of Metali… Amanda Krie… NA TRUE Metalinguis…
#> 2 W2371085312 Trastornos cognitivos… Valeria Abu… Aldo Ferre… FALSE Human immun…
#> 3 W2962551760 Conversations with a … Colwyn Trev… NA FALSE Action (phy…
#> 4 W2055502576 Preserved category le… Catherine H… Michael Co… FALSE Amnesia, Re…
#> 5 W2045105551 The Use of Profanity … John M. Rin… Po H. Lu FALSE Fluency, Ap…Additionally, opting out of the data frame conversion also means that
you can use your own preferred implementation for converting the list
output. This can be a very powerful optimization strategy when combined
with the select option.
For example, if you know that you are only selecting scalar fields,
you can very quickly convert the list output into a tidy data using more
powerful tools like data.table::rbindlist() or even just
rbind():
oa_fetch(
entity = "works",
topics.id = language_development$id,
options = list(sample = 5, seed = 1,
select = c("id", "display_name", "cited_by_count")),
output = "list"
) %>%
do.call(rbind.data.frame, .) %>%
as_tibble()
#> # A tibble: 5 × 3
#> id display_name cited_by_count
#> <chr> <chr> <int>
#> 1 https://openalex.org/W2969728018 Examination of Metalinguistic… 0
#> 2 https://openalex.org/W2371085312 Trastornos cognitivos en paci… 0
#> 3 https://openalex.org/W2962551760 Conversations with a two-mont… 101
#> 4 https://openalex.org/W2055502576 Preserved category learning i… 9
#> 5 https://openalex.org/W2045105551 The Use of Profanity During L… 12The oa_generate() strategy
If your code still seems slow, it is possible that you may have run
out of memory (especially when you do a snowball search like with
oa_snowball). In such cases, it might help to chunk your
work and save the output of each step, then piece them back together
later in a different session/program.1
The oa_generate() function is a lower-level function
that allows you to process one record at a time. This way, you can
process records in batches of, say, 1000 records, and write them out to
disk as you go along.2
In the example below, we show how oa_generate() works
when we want to find all the works that cite W1160808132.
query_url <- "https://api.openalex.org/works?filter=cites%3AW1160808132"
oar <- oa_generate(query_url, verbose = TRUE)
p1 <- oar() # record 1
#> Getting record 1 of 546 records...
p2 <- oar() # record 2
#> Getting record 2 of 546 records...
p3 <- oar() # record 3
#> Getting record 3 of 546 records...
head(p1)
#> $id
#> [1] "https://openalex.org/W2766937672"
#>
#> $doi
#> [1] "https://doi.org/10.1016/j.enpol.2017.10.050"
#>
#> $title
#> [1] "How economic growth, renewable electricity and natural resources contribute to CO2 emissions?"
#>
#> $display_name
#> [1] "How economic growth, renewable electricity and natural resources contribute to CO2 emissions?"
#>
#> $publication_year
#> [1] 2017
#>
#> $publication_date
#> [1] "2017-11-22"
head(p3)
#> $id
#> [1] "https://openalex.org/W2317269391"
#>
#> $doi
#> [1] "https://doi.org/10.1016/j.renene.2016.03.078"
#>
#> $title
#> [1] "Determinants of CO2 emissions in the European Union: The role of renewable and non-renewable energy"
#>
#> $display_name
#> [1] "Determinants of CO2 emissions in the European Union: The role of renewable and non-renewable energy"
#>
#> $publication_year
#> [1] 2016
#>
#> $publication_date
#> [1] "2016-04-01"As you see, each record returned by oa_generate is a
list of fields belonging to a work, parsed from the JSON response from
OpenAlex. You can process these records as you see fit, such as writing
them out as .rds files in batches of 100 records.
query_url <- "https://api.openalex.org/works?filter=cites%3AW1160808132"
oar <- oa_generate(query_url)
n <- 100
recs <- vector("list", n)
i <- 0
coro::loop(for (x in oar) {
j <- i %% n + 1
recs[[j]] <- x
if (j == n) {
saveRDS(recs, file.path(tempdir(), sprintf("rec-%s.rds", i %/% n)))
recs <- vector("list", n) # reset recs
}
i <- i + 1
})
dir(tempdir(), pattern = "rec-\\d.rds$")
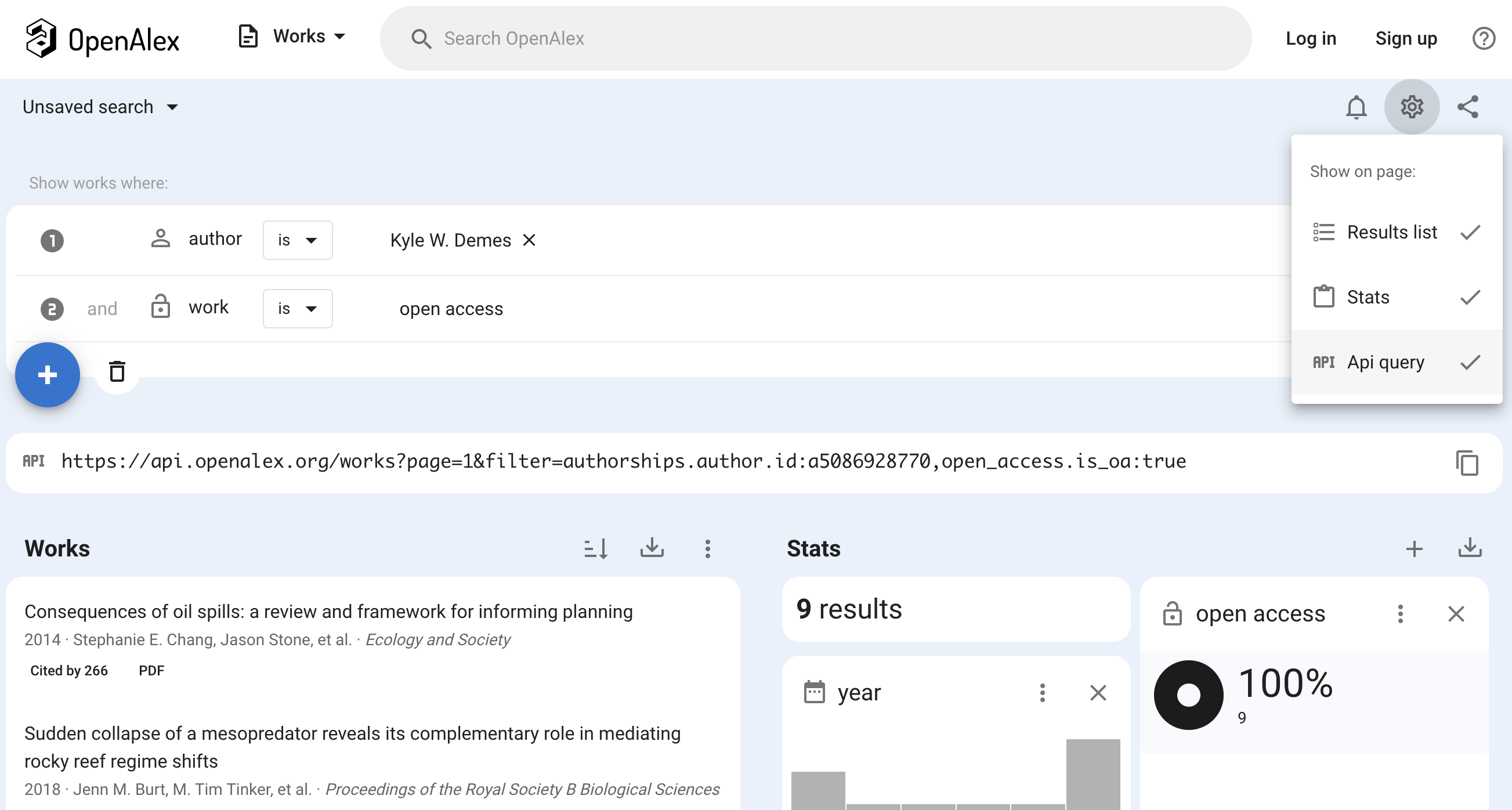
#> [1] "rec-0.rds" "rec-1.rds" "rec-2.rds" "rec-3.rds"Tips on generating the query URL to the OpenAlex API
To build your query, you can use oa_query() and
carefully read the API
documentation to see what fields/filters are available. For example,
I know cites is a filter we can use:
oa_query(entity = "works", cites = "W1160808132")
#> [1] "https://api.openalex.org/works?filter=cites%3AW1160808132"However, you might find it helpful to use the OpenAlex web interface to build the query interactively. Make sure you select the Gear icon on the right and toggle on “Api query”.