FedData is a package for downloading data from federated
(usually US Federal) data sets, using a consistent, spatial-first API.
In this vignette, you’ll learn how to load the FedData
package, download data for an “template” area of interest, and make
simple web maps using the mapview
package.
Load FedData and define a study area
# Load FedData and magrittr
library(FedData)
library(magrittr)
library(terra)
# Install mapview if necessary
if (!require("mapview")) {
install.packages("mapview")
}
library(mapview)
mapviewOptions(
basemaps = c(),
homebutton = FALSE,
query.position = "topright",
query.digits = 2,
query.prefix = "",
legend.pos = "bottomright",
platform = "leaflet",
fgb = TRUE,
georaster = TRUE
)
# Create a nice mapview template
plot_map <-
function(x, ...) {
if (inherits(x, "SpatRaster")) {
x %<>%
as("Raster")
}
bounds <-
FedData::meve %>%
sf::st_bbox() %>%
as.list()
mapview::mapview(x, ...)@map %>%
leaflet::removeLayersControl() %>%
leaflet::addTiles(
urlTemplate = "https://basemap.nationalmap.gov/ArcGIS/rest/services/USGSShadedReliefOnly/MapServer/tile/{z}/{y}/{x}"
) %>%
leaflet::addTiles(
urlTemplate = "https://tiles.stadiamaps.com/tiles/stamen_toner_lines/{z}/{x}/{y}.png"
) %>%
leaflet::addTiles(
urlTemplate = "https://tiles.stadiamaps.com/tiles/stamen_toner_labels/{z}/{x}/{y}.png"
) %>%
# leaflet::addProviderTiles("Stamen.TonerLines") %>%
# leaflet::addProviderTiles("Stamen.TonerLabels") %>%
leaflet::addPolygons(
data = FedData::meve,
color = "black",
fill = FALSE,
options = list(pointerEvents = "none"),
highlightOptions = list(sendToBack = TRUE)
) %>%
leaflet::fitBounds(
lng1 = bounds$xmin,
lng2 = bounds$xmax,
lat1 = bounds$ymin,
lat2 = bounds$ymax
)
}
# FedData comes loaded with the boundary of Mesa Verde National Park, for testing
FedData::meve
#> Geometry set for 1 feature
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: -108.5547 ymin: 37.15642 xmax: -108.3396 ymax: 37.35052
#> Geodetic CRS: WGS 84
plot_map(FedData::meve,
legend = FALSE
)Datasets
ORNL Daymet
# Get the DAYMET (North America only)
# Returns a raster
DAYMET <- get_daymet(
template = FedData::meve,
label = "meve",
elements = c("prcp", "tmax"),
years = 1985
)
plot_map(DAYMET$tmax$`1985-10-23`,
layer.name = "TMAX on 23 Oct 1985 (Degrees C)",
maxpixels = terra::ncell(DAYMET$tmax)
)NOAA GHCN-daily
# Get the daily GHCN data (GLOBAL)
# Returns a list: the first element is the spatial locations of stations,
# and the second is a list of the stations and their daily data
GHCN.prcp <- get_ghcn_daily(
template = FedData::meve,
label = "meve",
elements = c("prcp")
)
GHCN.prcp$spatial %>%
dplyr::mutate(label = paste0(ID, ": ", NAME)) %>%
plot_map(
label = "label",
layer.name = "GHCN_prcp",
legend = FALSE
)Standardized data
# Elements for which you require the same data
# (i.e., minimum and maximum temperature for the same days)
# can be standardized using standardize==T
GHCN.temp <- get_ghcn_daily(
template = FedData::meve,
label = "meve",
elements = c("tmin", "tmax"),
years = 1980:1985,
standardize = TRUE
)
GHCN.temp$spatial %>%
dplyr::mutate(label = paste0(ID, ": ", NAME)) %>%
plot_map(
label = "label",
layer.name = "GHCN_temp",
legend = FALSE
)USGS National Hydrography Dataset
# Get the NHD (USA ONLY)
NHD <-
get_nhd(
template = FedData::meve,
label = "meve",
force.redo = TRUE
)
# FedData comes with a simple plotting function for NHD data
plot_nhd(NHD, template = FedData::meve)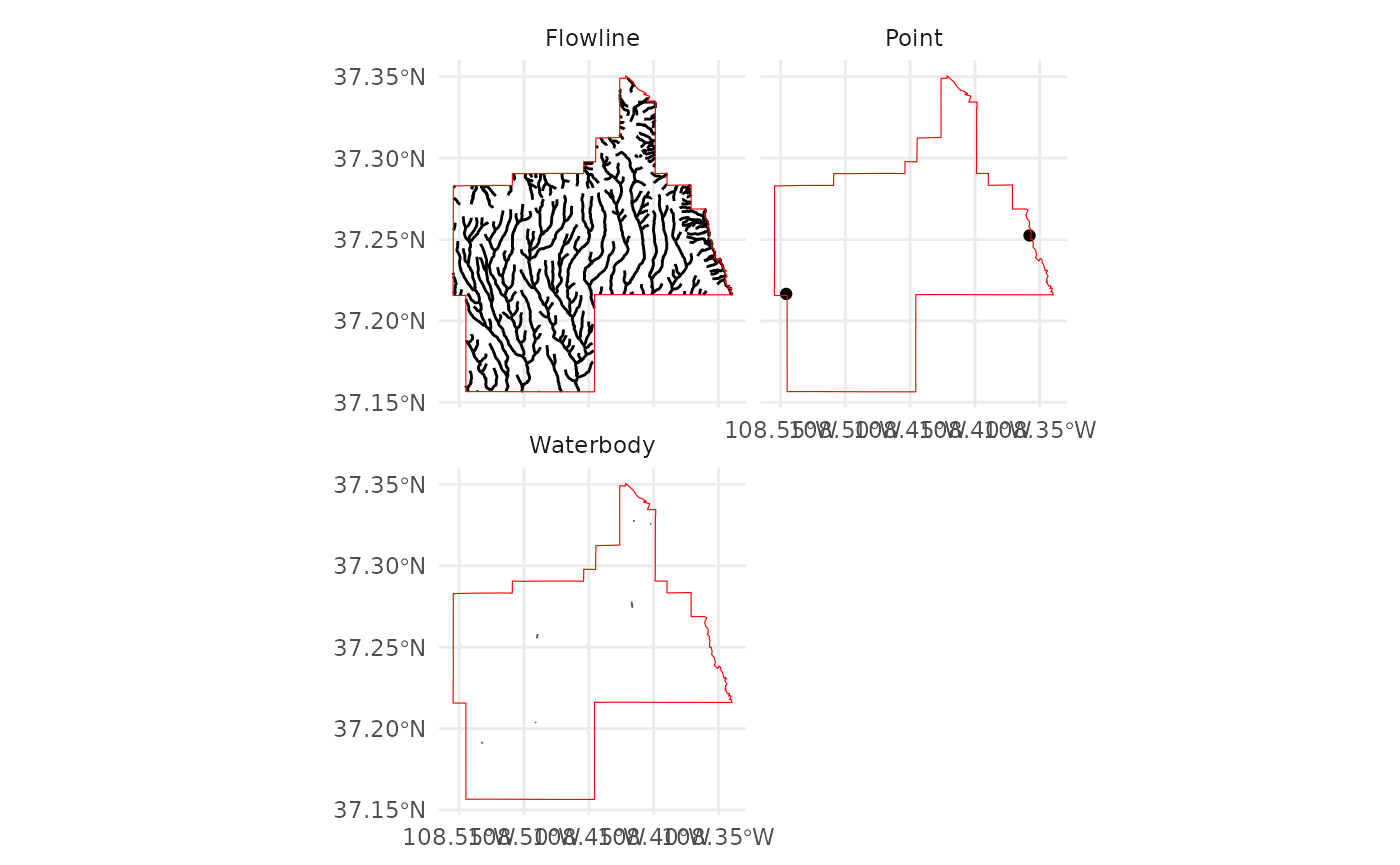
USDA NRCS SSURGO
# Get the NRCS SSURGO data (USA ONLY)
SSURGO.MEVE <- get_ssurgo(
template = FedData::meve,
label = "meve"
)
SSURGO.MEVE$spatial %>%
rmapshaper::ms_simplify() %>%
dplyr::left_join(
SSURGO.MEVE$tabular$mapunit %>%
dplyr::mutate(mukey = as.character(mukey)),
by = c("MUKEY" = "mukey")
) %>%
dplyr::mutate(label = paste0(MUKEY, ": ", muname)) %>%
dplyr::select(-musym) %>%
plot_map(
zcol = "label",
legend = FALSE
)SSURGO by Soil Survey Area
# Or, download by Soil Survey Area names
SSURGO.areas <- get_ssurgo(
template = c("CO670"),
label = "CO670"
)
SSURGO.areas$spatial %>%
rmapshaper::ms_simplify() %>%
dplyr::left_join(
SSURGO.areas$tabular$muaggatt %>%
dplyr::mutate(mukey = as.character(mukey)),
by = c("MUKEY" = "mukey")
) %>%
dplyr::mutate(label = paste0(MUKEY, ": ", muname)) %>%
dplyr::select(-musym) %>%
plot_map(
zcol = "brockdepmin",
layer.name = "Minimum Bedrock Depth (cm)",
label = "label"
)ITRDB
# Get the ITRDB records
# Buffer MEVE, because there aren't any chronologies in the Park
ITRDB <-
get_itrdb(
template = FedData::meve %>%
sf::st_buffer(50000),
label = "meve",
measurement.type = "Ring Width",
chronology.type = "Standard"
)
#> Warning: attribute variables are assumed to be spatially constant throughout
#> all geometries
plot_map(ITRDB$metadata,
zcol = "SPECIES",
layer.name = "Species",
label = "NAME"
) %>%
leaflet::setView(
lat = sf::st_centroid(FedData::meve) %>%
sf::st_coordinates() %>%
as.numeric() %>%
magrittr::extract2(2),
lng = sf::st_centroid(FedData::meve) %>%
sf::st_coordinates() %>%
as.numeric() %>%
magrittr::extract2(1),
zoom = 9
)USGS National Land Cover Dataset
# Get the NLCD (USA ONLY)
# Returns a raster
NLCD <- get_nlcd(
template = FedData::meve,
year = 2019,
label = "meve"
)
plot(NLCD)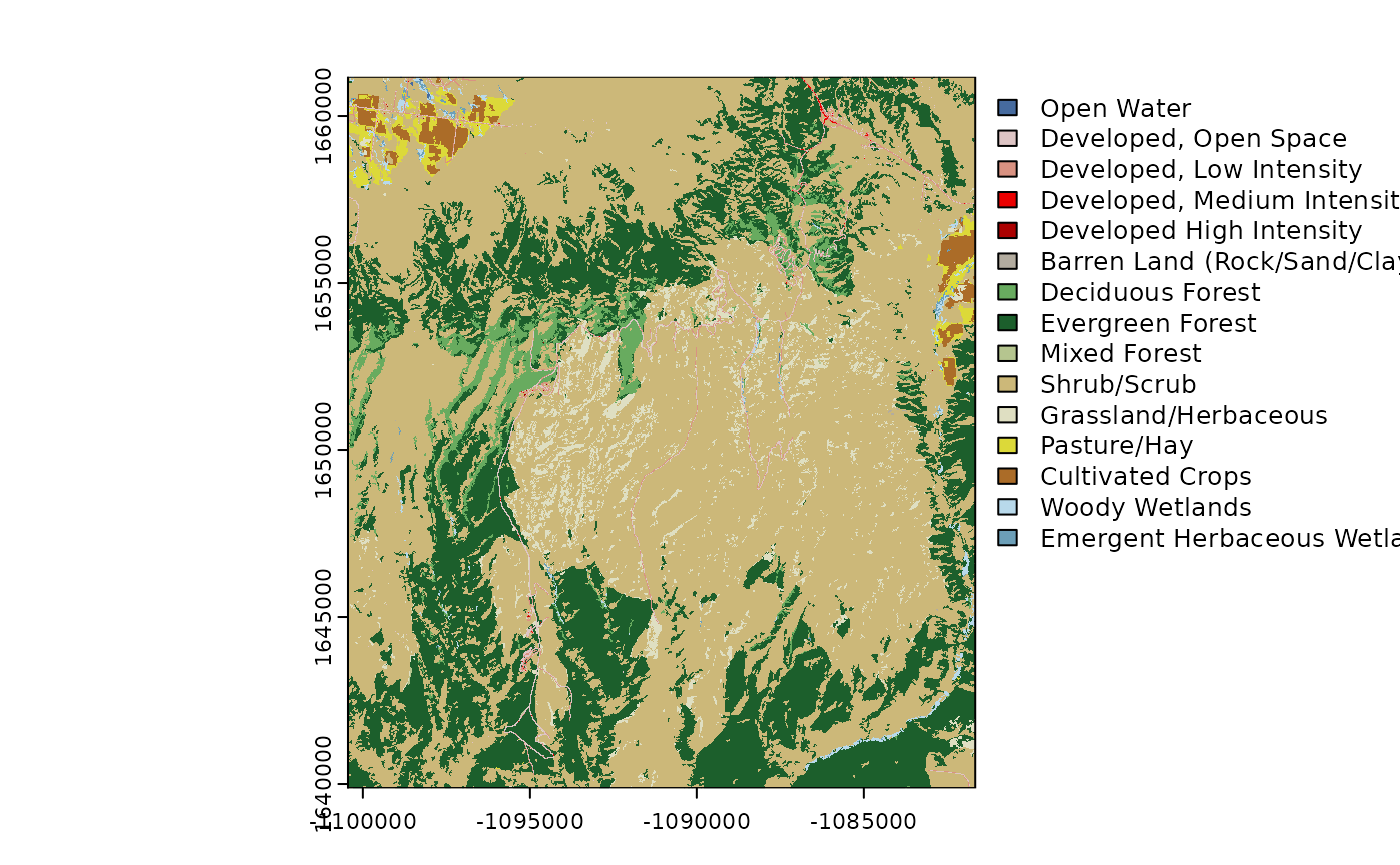
# Also get the impervious or canopy datasets
NLCD_impervious <-
get_nlcd(
template = FedData::meve,
year = 2019,
label = "meve_impervious",
dataset = "impervious"
)
plot(NLCD_impervious)
NLCD_canopy <-
get_nlcd(
template = FedData::meve,
year = 2019,
label = "meve_canopy",
dataset = "canopy"
)
plot(NLCD_canopy)
Annual National Landcover Dataset
# Get the NLCD (USA ONLY)
# Returns a raster
NLCD_ANNUAL <-
get_nlcd_annual(
template = FedData::meve,
label = "meve",
year = 2020,
product =
c(
"LndCov",
"LndChg",
"LndCnf",
"FctImp",
"ImpDsc",
"SpcChg"
)
)
# Returns a data.frame of all downloaded data
NLCD_ANNUAL
#> # A tibble: 6 × 7
#> # Rowwise:
#> product year region collection version outfile rast
#> <ord> <int> <ord> <int> <int> <glue> <list>
#> 1 LndCov 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
#> 2 LndChg 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
#> 3 LndCnf 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
#> 4 FctImp 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
#> 5 ImpDsc 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
#> 6 SpcChg 2020 CU 1 0 /tmp/RtmpGBja35/Fe… <SpatRstr[,627,1]>
plot(NLCD_ANNUAL$rast[[1]])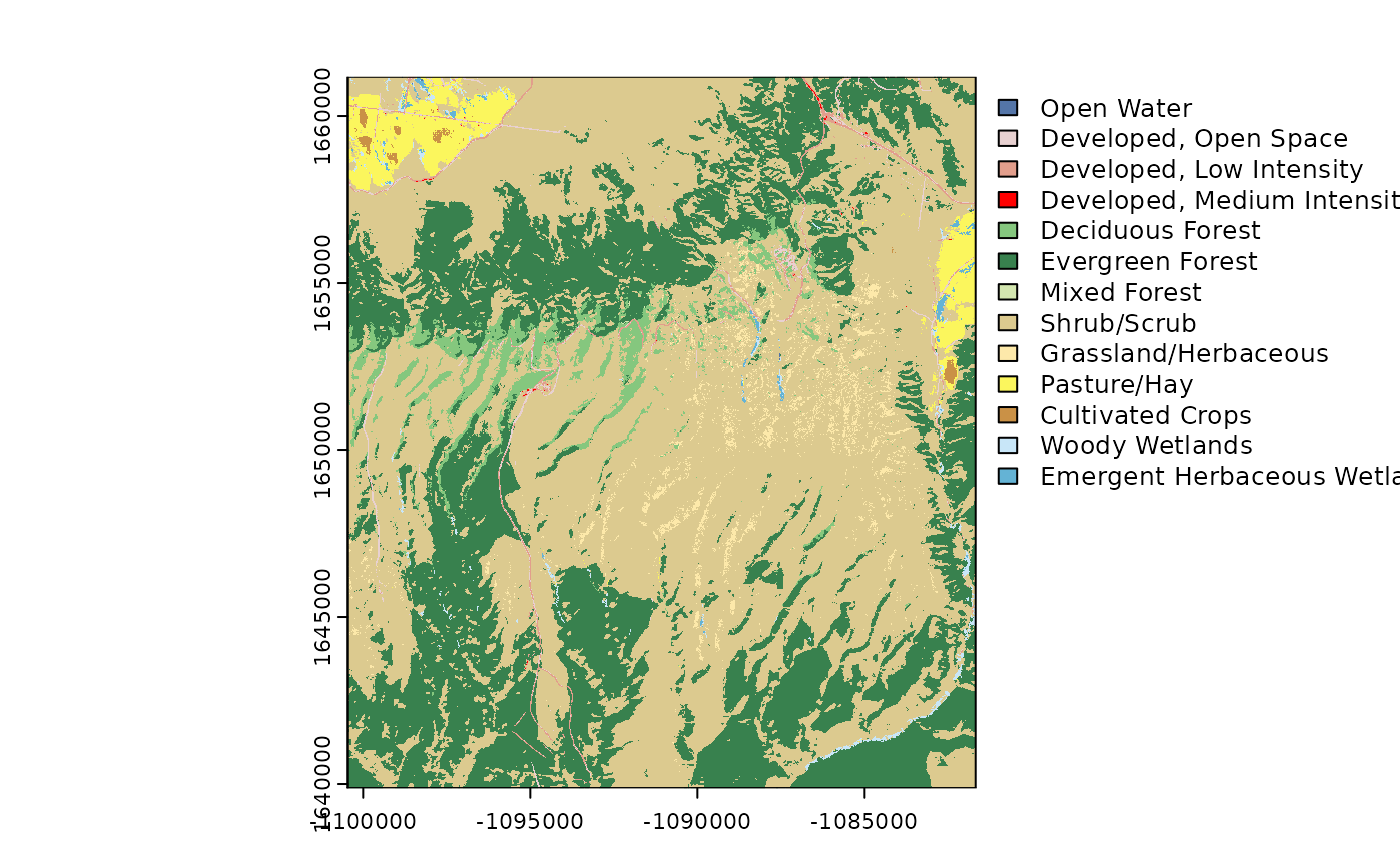
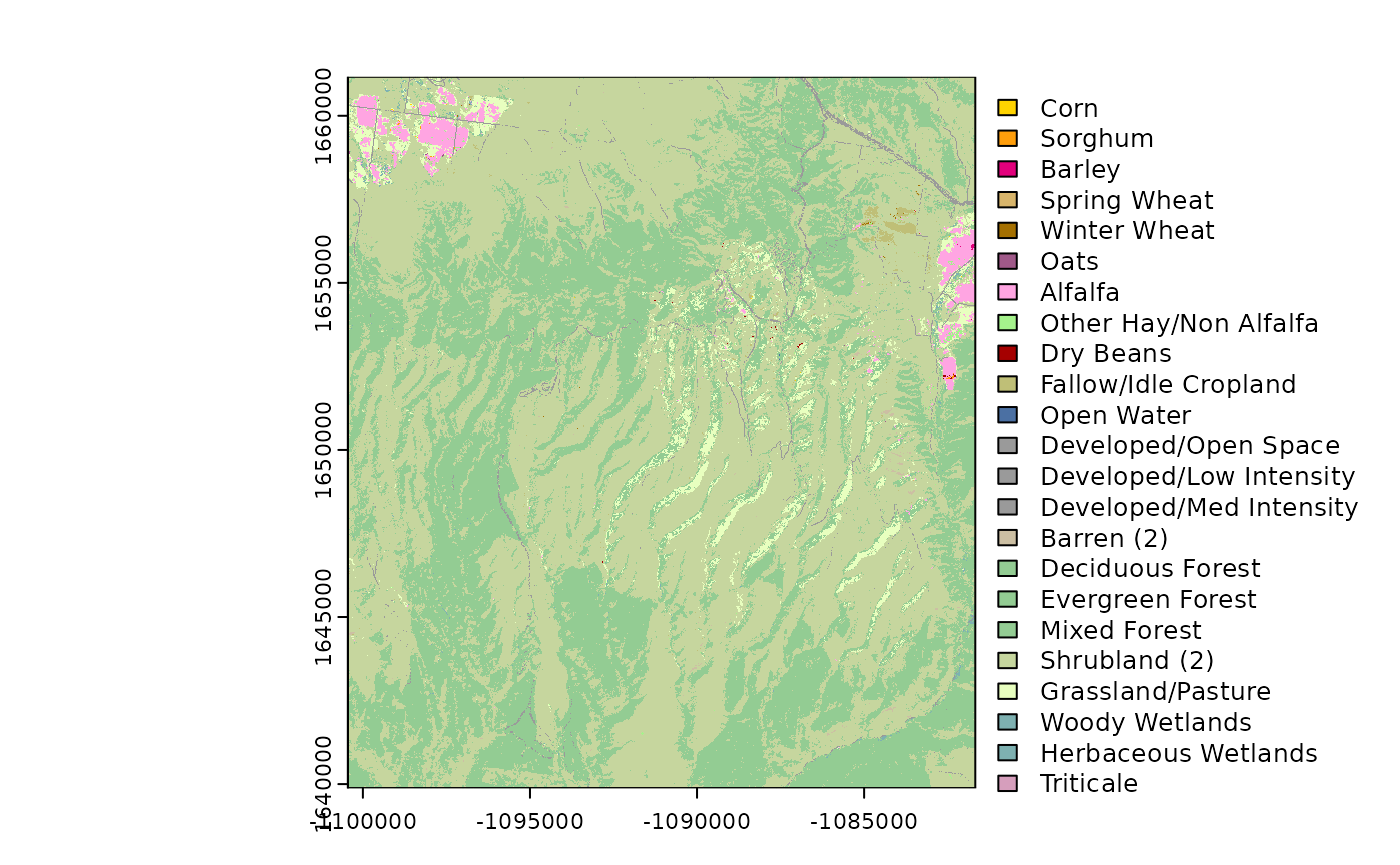
USDA NASS Cropland Data Layer
# Get the NASS (USA ONLY)
# Returns a raster
NASS_CDL <- get_nass_cdl(
template = FedData::meve,
year = 2016,
label = "meve"
)
plot(NASS_CDL)