Main function for the MODIS Time Series Processing Tool (MODIStsp)
Usage
MODIStsp(
...,
gui = TRUE,
out_folder = NULL,
out_folder_mod = NULL,
opts_file = NULL,
selprod = NULL,
prod_version = NULL,
bandsel = NULL,
quality_bandsel = NULL,
indexes_bandsel = NULL,
sensor = NULL,
download_server = NULL,
downloader = NULL,
user = NULL,
password = NULL,
download_range = NULL,
start_date = NULL,
end_date = NULL,
spatmeth = NULL,
start_x = NULL,
end_x = NULL,
start_y = NULL,
end_y = NULL,
bbox = NULL,
spafile = NULL,
out_projsel = NULL,
output_proj = NULL,
out_res_sel = NULL,
out_res = NULL,
resampling = NULL,
reprocess = NULL,
delete_hdf = NULL,
nodata_change = NULL,
scale_val = NULL,
ts_format = NULL,
out_format = NULL,
compress = NULL,
test = NULL,
n_retries = 5,
verbose = TRUE,
parallel = TRUE
)Arguments
- ...
not used for values, forces later arguments to bind by name
- gui
logicalif TRUE: the GUI is opened before processing. If FALSE: processing parameters are retrieved from the providedopts_fileargument), Default: TRUE- out_folder
characterMain output folder, default: NULL.- out_folder_mod
characterOutput folder for original HDF storage. If"$tempdir"(default), a temporary directory is used.- opts_file
characterfull path to a JSON file containing MODIStsp processing options saved from the GUI, Default: NULL- selprod
characterName of selected MODIS product (e.g., Vegetation Indexes_16Days_250m (M*D13Q1)). You can get a list of available product names using functionMODIStsp_get_prodnames, Default: NULL- prod_version
Version of the selected MODIS product. Currently versions
"061"and/or"006"can be chosen. Default value is"061"(version"006"was decommissioned by USGS on July 31, 2023, products of this version are being gradually removed). In case users would encounter an error in the encoding of bands or quality flags they are encouraged to report it by opening a new issue on GitHub at https://github.com/ropensci/MODIStsp/issues.- bandsel
character arrayOriginal MODIS layers to be processed. You can get a list of available layers for a given product using functionMODIStsp_get_prodlayers(e.g., MODIStsp_get_prodlayers("M*D13Q1")$bandnames), Default: NULL- quality_bandsel
character arrayQuality Indicators to be computed starting from bit fields of original MODIS layers. You can get a list of available quality layers for a given product using functionMODIStsp_get_prodlayers(e.g., MODIStsp_get_prodlayers("M*D13Q1")$quality_bandnames), Default: NULL- indexes_bandsel
character arraySpectral Indexes to be computed starting from reflectance bands. You can get a list of available quality layers for a given product using functionMODIStsp_get_prodlayers(e.g., MODIStsp_get_prodlayers("M*D13Q1")$indexes_bandnames), Default: NULL- sensor
character ["Terra"| "Aqua" | "Both"]MODIS platform to be considered. (Ignored for MCD* products). Default: "Both"- download_server
character ["http" | "offline"]service to be used for download. Default: "http"- downloader
download_server
character ["http" | "aria2"]downloader to be used, Default: "http"- user
characterUsername for NASA http server. (urs.earthdata.nasa.gov/home).- password
characterPassword for NASA http server (urs.earthdata.nasa.gov/home).- download_range
character ["Full" | "Seasonal"]If "full", all the available images between the starting and the ending dates are downloaded; If "seasonal", only the images included in the season are downloaded (e.g: if the starting date is 2005-12-01 and the ending is 2010-02-31, only the images of December, January and February from 2005 to 2010 - excluding 2005-01, 2005-02 and 2010-12 - are downloaded), Default: Full- start_date
characterStart date for images download and preprocessing (yyyy.mm.dd), Default: NULL- end_date
characterEnd date for images download and preprocessing (yyyy.mm.dd), Default: NULL- spatmeth
character ["tiles" | "bbox" | "file"], indicates how the processing extent is retrieves. if "tiles", use the specified tiles (start_x....). If "file", retrieve extent from spatial file specifies inspafile. If "bbox", use the specified bounding box, Default: "tiles"- start_x
integer [0-35]Start MODIS horizontal tile defining spatial extent. Ignored if spatmeth != "tiles", Default: 18- end_x
integer [0-35]End MODIS horizontal tile defining spatial extent. Ignored if spatmeth != "tiles", Default: 18- start_y
integer [0-17]Start MODIS vertical tile defining spatial extent. Ignored if spatmeth != "tiles", Default: 4- end_y
integer [0-17]End MODIS vertical tile defining spatial extent. Ignored if spatmeth != "tiles", Default: 4- bbox
numeric(4)Output bounding box (xmin, ymin, xmax, ymax) in out_proj coordinate system. Ignored if spatmeth == "tiles", Default: NULL- spafile
character(optional) full path of a spatial file to use to derive the processing extent. If not NULL, the processing options which define the extent, the selected tiles and the "Full Tile / Custom" in the JSON options file are overwritten and new files are created on the extent of the provided spatial file. Ignored if spatmeth != "file", Default: NULL- out_projsel
character ["Native", "User Defined] If "Native", the outputs keep the original resolution of MODIS HDF images. Otherwise, the value set in "out_res" is used, Default:Native- output_proj
charactereither equal to "MODIS Sinusoidal", or to the code of a valid EPSG or to a WKT projection string. Ignored if outproj_sel == "Native", Default: NULL- out_res_sel
character ["Native", "User Defined]. If "Native", the outputs keep the original resolution of MODIS HDF images. Otherwise, the value set in "out_res" is used.- out_res
floatOutput resolution (in output projection measurement unit). Ignored if out_res_sel == "Native".- resampling
character ["near" | "bilinear" | "cubic" | "cubicspline", |lanczos"|, "average"|, "mode", |"max"|, |"min"|, |"q1"|, |"q3"|, |"sum"|]Resampling method to be used bygdalwarp.- reprocess
logicalIf TRUE, reprocess data for already existing dates.- delete_hdf
logicalIf TRUE, delete downloaded HDF files after completion.- nodata_change
logicalif TRUE, NoData values are set to the max value of the datatype of the layer on the MODIStsp output rasters. NOTE: If multiple nodata values are reported for a layer, all are reset to the new value.- scale_val
logicalIf TRUE, scale and offset are applied to original MODIS layers, and Spectral Indexes are saved as floating point. If FALSE, no rescaling is done and Spectral Indexes are saved as integer, with a 10000 scaling factor.- ts_format
character array including ["R RasterStack" | "ENVI Meta Files" | "GDAL VRT" | "ENVI and GDAL"]Selected virtual time series format.- out_format
character ["ENVI" | "GTiff"]Desired output format.- compress
character ["None" | "PACKBITS" | "LZW" | "DEFLATE"]Compression method for GTiff outputs (Ignored ifout_format == ENVI)- test
integer | character (e.g., "01a")if set, MODIStsp is executed in "test mode", using a preset Options File instead than opening the GUI or accepting theopts_fileparameter. This allows both to check correct installation on user's machines, and to implement unit testing.- n_retries
numericmaximum number of retries on download functions. In case any download function fails more thann_retriestimes consecutively, MODIStsp_process will abort, Default: 20- verbose
logicalIf FALSE, suppress processing messages, Default: TRUE- parallel
logicalIf TRUE (default), the function is run using parallel processing, to speed-up the computation for large rasters (with a maximum of 8 cores). The number of cores is automatically determined; specifying it is also possible (e.g.parallel = 4). In this case, more than 8 cores can be specified. If FALSE (default), single core processing is used.
Details
The function is used to:
initialize the processing (folder names, packages, etc.);
launch the GUI (
MODIStsp_GUI()) on interactive execution, or load an options file to set processing arguments and/or retrieve CLI inputs and run processing on non-interactive execution;launch the routines for downloading and processing the requested datasets. (
MODIStsp_process())launching the function with GUI = FALSE and without specifying a opts_file initializes arguments with default values. This allows making a test run.
Examples
# \donttest{
#' # - Running the tool using the GUI
# Running the tool without any option will start the GUI with the default or
# last used settings, in interactive mode (i.e., with gui = TRUE).
if (interactive()) {
MODIStsp()
}
#' # - Running the tool specifying processing arguments in the call
# **NOTE** Output files of examples are saved to file.path(tempdir(), "MODIStsp").
# Here we process layers __NDVI__ and __EVI__ and quality indicator __usefulness__
# of product __M*D13Q1__, considering both Terra and Aqua platforms, for dates
# comprised between 2020-06-01 and 2020-06-15 and saves output to R tempdir
# --> See name and available layers for product M*D13Q1.
# Note that this example (as well as the following ones) is run in single
# core to follow CRAN policies, by setting parallel = FALSE.
# Users can exploit multicore functionalities skipping to set this argument.
# The following check is performed in order not to provide errors
# running the examples if HDF4 is not supported.
is_hdf4_supported <- "HDF4" %in% sf::st_drivers("raster")$name
MODIStsp_get_prodlayers("M*D13A2")
#> $prodname
#> [1] "Vegetation_Indexes_16Days_1Km (M*D13A2)"
#>
#> $bandnames
#> [1] "NDVI" "EVI" "VI_QA" "b1_Red" "b2_NIR" "b3_Blue"
#> [7] "b7_SWIR" "View_Zen" "Sun_Zen" "Rel_Az" "DOY" "Rely"
#>
#> $bandfullnames
#> [1] "16 day NDVI average" "16 day EVI average"
#> [3] "VI quality indicators" "Surface Reflectance Band 1"
#> [5] "Surface Reflectance Band 2" "Surface Reflectance Band 3"
#> [7] "Surface Reflectance Band 7" "View zenith angle of VI pixel"
#> [9] "Sun zenith angle of VI pixel" "Relative azimuth angle of VI pixel"
#> [11] "Day of year of VI pixel" "Quality reliability of VI pixel"
#>
#> $quality_bandnames
#> [1] "QA_qual" "QA_usef" "QA_aer" "QA_adj_cld" "QA_BRDF"
#> [6] "QA_mix_cld" "QA_land_wat" "QA_snow_ice" "QA_shd"
#>
#> $quality_fullnames
#> [1] "VI Quality"
#> [2] "VI usefulness"
#> [3] "Aerosol quantity"
#> [4] "Adjacent cloud detected"
#> [5] "Atmosphere BRDF correction performed"
#> [6] "Mixed Clouds"
#> [7] "Land/Water Flag"
#> [8] "Possible snow/ice"
#> [9] "Possible shadow"
#>
#> $indexes_bandnames
#> [1] "SR" "NDFI" "NDII7" "SAVI"
#>
#> $indexes_fullnames
#> [1] "Simple Ratio (NIR/Red)"
#> [2] "Flood Index (Red-SWIR2)/(Red+SWIR2)"
#> [3] "NDII7 (NIR-SWIR2)/(NIR+SWIR2)"
#> [4] "SAVI (NIR-Red)/(NIR+Red+0.5)*(1+0.5)"
#>
if (is_hdf4_supported) {
MODIStsp(
gui = FALSE,
out_folder = "$tempdir",
selprod = "Vegetation_Indexes_16Days_1Km (M*D13A2)",
bandsel = c("EVI", "NDVI"),
quality_bandsel = "QA_usef",
indexes_bandsel = "SR",
user = "mstp_test" ,
password = "MSTP_test_01",
start_date = "2020.06.01",
end_date = "2020.06.15",
verbose = FALSE,
parallel = FALSE
)
}
#' # - Running the tool using the settings previously saved in a specific options file
# **NOTE** Output files of examples are saved to file.path(tempdir(), "MODIStsp").
# You can run the examples with `gui = TRUE` to set a different output folder!
# Here we use a test json file saved in MODIStsp installation folder which
# downloads and processed 3 MOD13A2 images over the Como Lake (Lombardy, Italy)
# and retrieves NDVI and EVI data, plus the Usefulness Index Quality Indicator.
opts_file <- system.file("testdata/test_MOD13A2.json", package = "MODIStsp")
if (is_hdf4_supported) {
MODIStsp(gui = FALSE, opts_file = opts_file, verbose = TRUE, parallel = FALSE)
}
#> GDAL version in use: 3.4.1
#> [Sat Feb 10 06:25:47 2024] MODIStsp --> Starting processing
#> [Sat Feb 10 06:25:47 2024] Accessing http server at: https://e4ftl01.cr.usgs.gov/MOLT/MOD13A2.061/
#> [Sat Feb 10 06:25:48 2024] Retrieving list of available ` Terra ` Files for Year 2016
#> [Sat Feb 10 06:25:49 2024] Downloading Terra Files for date: 2016_06_09 : 1 of: 1
#> [Sat Feb 10 06:25:50 2024] [Sat Feb 10 06:25:50 2024] 1 files for date: 2016.06.09 were successfully downloaded!
#> [Sat Feb 10 06:25:50 2024] Processing Terra NDVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:50 2024] Processing Terra EVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:50 2024] Processing Terra VI_QA files for date: 2016_06_09
#> [Sat Feb 10 06:25:50 2024] Computing QA_usef for date: 2016_06_09
#> [Sat Feb 10 06:25:52 2024] Downloading Terra Files for date: 2016_06_25 : 1 of: 1
#> [Sat Feb 10 06:25:52 2024] [Sat Feb 10 06:25:52 2024] 1 files for date: 2016.06.25 were successfully downloaded!
#> [Sat Feb 10 06:25:52 2024] Processing Terra NDVI files for date: 2016_06_25
#> [Sat Feb 10 06:25:53 2024] Processing Terra EVI files for date: 2016_06_25
#> [Sat Feb 10 06:25:53 2024] Processing Terra VI_QA files for date: 2016_06_25
#> [Sat Feb 10 06:25:53 2024] Computing QA_usef for date: 2016_06_25
#> [Sat Feb 10 06:25:53 2024] Creating Virtual Files and R time series for layer NDVI
#> [Sat Feb 10 06:25:53 2024] Creating Virtual Files and R time series for layer EVI
#> [Sat Feb 10 06:25:53 2024] Creating Virtual Files and R time series for layer QA_usef
#> [Sat Feb 10 06:25:53 2024] Total Processing Time: 6.12111711502075
#> [Sat Feb 10 06:25:53 2024] MODIStsp processed files are in: `/tmp/RtmprleXzN/MODIStsp`
#> [Sat Feb 10 06:25:53 2024] Original downloaded MODIS HDF files are in: `/tmp/RtmprleXzN/MODIStsp/HDFs`
#> [Sat Feb 10 06:25:53 2024] Processing options saved to: `/tmp/RtmprleXzN/MODIStsp/MODIStsp_2024-02-10.json`
# Running the tool using the settings previously saved in a specific option file
# and specifying the extent from a spatial file allows to re-use the same
# processing settings to perform download and reprocessing on a different area
opts_file <- system.file("testdata/test_MOD13A2.json", package = "MODIStsp")
spatial_file <- system.file("testdata/lakeshapes/garda_lake.shp", package = "MODIStsp")
if (is_hdf4_supported) {
MODIStsp(
gui = FALSE,
opts_file = opts_file,
spatmeth = "file",
spafile = spatial_file,
verbose = TRUE,
parallel = FALSE
)
}
#> GDAL version in use: 3.4.1
#> [Sat Feb 10 06:25:53 2024] MODIStsp --> Starting processing
#> [Sat Feb 10 06:25:53 2024] Accessing http server at: https://e4ftl01.cr.usgs.gov/MOLT/MOD13A2.061/
#> [Sat Feb 10 06:25:54 2024] Retrieving list of available ` Terra ` Files for Year 2016
#> [Sat Feb 10 06:25:54 2024] HDF File: MOD13A2.A2016161.h18v04.061.2021353012725.hdf already exists on your system. Skipping download!
#> [Sat Feb 10 06:25:54 2024] [Sat Feb 10 06:25:54 2024] 1 files for date: 2016.06.09 were successfully downloaded!
#> [Sat Feb 10 06:25:55 2024] Processing Terra NDVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:55 2024] Processing Terra EVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:55 2024] Processing Terra VI_QA files for date: 2016_06_09
#> [Sat Feb 10 06:25:55 2024] Computing QA_usef for date: 2016_06_09
#> [Sat Feb 10 06:25:56 2024] HDF File: MOD13A2.A2016177.h18v04.061.2021353092234.hdf already exists on your system. Skipping download!
#> [Sat Feb 10 06:25:56 2024] [Sat Feb 10 06:25:56 2024] 1 files for date: 2016.06.25 were successfully downloaded!
#> [Sat Feb 10 06:25:56 2024] Processing Terra NDVI files for date: 2016_06_25
#> [Sat Feb 10 06:25:56 2024] Processing Terra EVI files for date: 2016_06_25
#> [Sat Feb 10 06:25:56 2024] Processing Terra VI_QA files for date: 2016_06_25
#> [Sat Feb 10 06:25:57 2024] Computing QA_usef for date: 2016_06_25
#> [Sat Feb 10 06:25:57 2024] Creating Virtual Files and R time series for layer NDVI
#> [Sat Feb 10 06:25:57 2024] Creating Virtual Files and R time series for layer EVI
#> [Sat Feb 10 06:25:57 2024] Creating Virtual Files and R time series for layer QA_usef
#> [Sat Feb 10 06:25:57 2024] Total Processing Time: 3.63410425186157
#> [Sat Feb 10 06:25:57 2024] MODIStsp processed files are in: `/tmp/RtmprleXzN/MODIStsp/garda_lake`
#> [Sat Feb 10 06:25:57 2024] Original downloaded MODIS HDF files are in: `/tmp/RtmprleXzN/MODIStsp/HDFs`
#> [Sat Feb 10 06:25:57 2024] Processing options saved to: `/tmp/RtmprleXzN/MODIStsp/garda_lake/MODIStsp_2024-02-10.json`
# Running the tool using the settings previously saved in a
# specific options file and specifying each time the extent from a different
# spatial file (e.g., to perform the same processing on several extents)
# Note that you can also put all your extent files in a specific folder and
# create the extent list using for example.
extent_list = list.files(
system.file("testdata/lakeshapes/", package = "MODIStsp"),
"\\.shp$",
full.names = TRUE
)
extent_list
#> [1] "/usr/local/lib/R/site-library/MODIStsp/testdata/lakeshapes//garda_lake.shp"
#> [2] "/usr/local/lib/R/site-library/MODIStsp/testdata/lakeshapes//iseo_lake.shp"
opts_file <- system.file("testdata/test_MOD13A2.json", package = "MODIStsp")
if (is_hdf4_supported) {
for (single_shape in extent_list) {
MODIStsp(
gui = FALSE,
opts_file = opts_file,
spatmeth = "file",
spafile = single_shape,
verbose = TRUE,
parallel = FALSE
)
}
}
#> GDAL version in use: 3.4.1
#> [Sat Feb 10 06:25:57 2024] MODIStsp --> Starting processing
#> [Sat Feb 10 06:25:57 2024] Accessing http server at: https://e4ftl01.cr.usgs.gov/MOLT/MOD13A2.061/
#> [Sat Feb 10 06:25:57 2024] Retrieving list of available ` Terra ` Files for Year 2016
#> [Sat Feb 10 06:25:57 2024] [Sat Feb 10 06:25:57 2024] All Required output files for date 2016_06_09 are already existing - Doing Nothing!
#> Set Reprocess to TRUE to reprocess existing data!
#> [Sat Feb 10 06:25:57 2024] [Sat Feb 10 06:25:57 2024] All Required output files for date 2016_06_25 are already existing - Doing Nothing!
#> Set Reprocess to TRUE to reprocess existing data!
#> [Sat Feb 10 06:25:57 2024] Creating Virtual Files and R time series for layer NDVI
#> [Sat Feb 10 06:25:58 2024] Creating Virtual Files and R time series for layer EVI
#> [Sat Feb 10 06:25:58 2024] Creating Virtual Files and R time series for layer QA_usef
#> [Sat Feb 10 06:25:58 2024] Total Processing Time: 0.557390213012695
#> [Sat Feb 10 06:25:58 2024] MODIStsp processed files are in: `/tmp/RtmprleXzN/MODIStsp/garda_lake`
#> [Sat Feb 10 06:25:58 2024] Original downloaded MODIS HDF files are in: `/tmp/RtmprleXzN/MODIStsp/HDFs`
#> [Sat Feb 10 06:25:58 2024] Processing options saved to: `/tmp/RtmprleXzN/MODIStsp/garda_lake/MODIStsp_2024-02-10.json`
#> GDAL version in use: 3.4.1
#> [Sat Feb 10 06:25:58 2024] MODIStsp --> Starting processing
#> [Sat Feb 10 06:25:58 2024] Accessing http server at: https://e4ftl01.cr.usgs.gov/MOLT/MOD13A2.061/
#> [Sat Feb 10 06:25:58 2024] Retrieving list of available ` Terra ` Files for Year 2016
#> [Sat Feb 10 06:25:59 2024] HDF File: MOD13A2.A2016161.h18v04.061.2021353012725.hdf already exists on your system. Skipping download!
#> [Sat Feb 10 06:25:59 2024] [Sat Feb 10 06:25:59 2024] 1 files for date: 2016.06.09 were successfully downloaded!
#> [Sat Feb 10 06:25:59 2024] Processing Terra NDVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:59 2024] Processing Terra EVI files for date: 2016_06_09
#> [Sat Feb 10 06:25:59 2024] Processing Terra VI_QA files for date: 2016_06_09
#> [Sat Feb 10 06:25:59 2024] Computing QA_usef for date: 2016_06_09
#> [Sat Feb 10 06:26:00 2024] HDF File: MOD13A2.A2016177.h18v04.061.2021353092234.hdf already exists on your system. Skipping download!
#> [Sat Feb 10 06:26:00 2024] [Sat Feb 10 06:26:00 2024] 1 files for date: 2016.06.25 were successfully downloaded!
#> [Sat Feb 10 06:26:00 2024] Processing Terra NDVI files for date: 2016_06_25
#> [Sat Feb 10 06:26:00 2024] Processing Terra EVI files for date: 2016_06_25
#> [Sat Feb 10 06:26:00 2024] Processing Terra VI_QA files for date: 2016_06_25
#> [Sat Feb 10 06:26:01 2024] Computing QA_usef for date: 2016_06_25
#> [Sat Feb 10 06:26:01 2024] Creating Virtual Files and R time series for layer NDVI
#> [Sat Feb 10 06:26:01 2024] Creating Virtual Files and R time series for layer EVI
#> [Sat Feb 10 06:26:01 2024] Creating Virtual Files and R time series for layer QA_usef
#> [Sat Feb 10 06:26:01 2024] Total Processing Time: 3.0306179523468
#> [Sat Feb 10 06:26:01 2024] MODIStsp processed files are in: `/tmp/RtmprleXzN/MODIStsp/iseo_lake`
#> [Sat Feb 10 06:26:01 2024] Original downloaded MODIS HDF files are in: `/tmp/RtmprleXzN/MODIStsp/HDFs`
#> [Sat Feb 10 06:26:01 2024] Processing options saved to: `/tmp/RtmprleXzN/MODIStsp/iseo_lake/MODIStsp_2024-02-10.json`
# output files are placed in separate folders:
outfiles_garda <- list.files(
file.path(tempdir(), "MODIStsp/garda_lake/VI_16Days_1Km_v61/NDVI"),
full.names = TRUE
)
outfiles_garda
#> [1] "/tmp/RtmprleXzN/MODIStsp/garda_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_161.tif"
#> [2] "/tmp/RtmprleXzN/MODIStsp/garda_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_177.tif"
require(raster)
#> Loading required package: raster
#> Loading required package: sp
if (length(outfiles_garda) > 0) {
plot(raster(outfiles_garda[1] ))
}
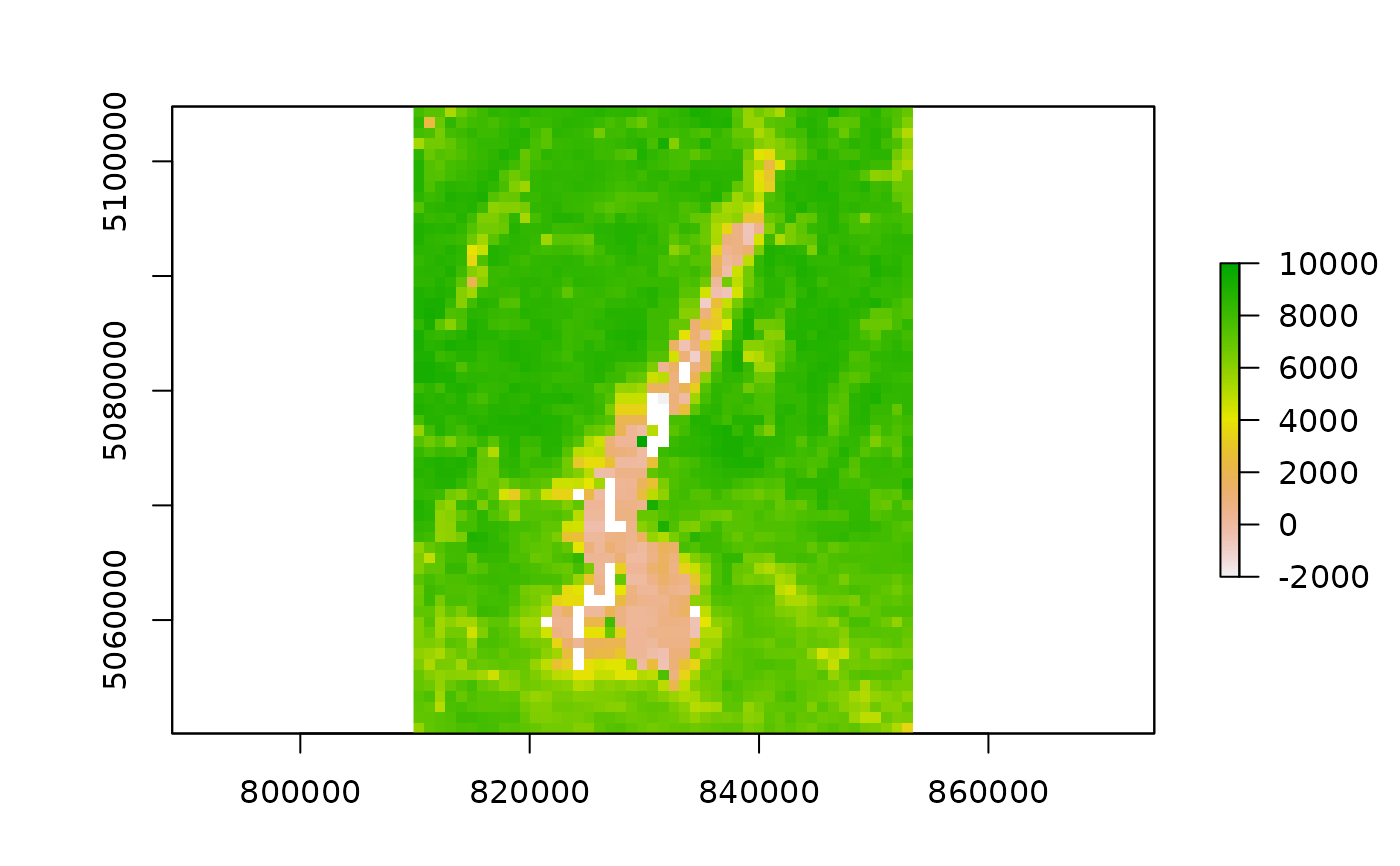 outfiles_iseo <- list.files(
file.path(tempdir(), "MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI"),
full.names = TRUE
)
outfiles_iseo
#> [1] "/tmp/RtmprleXzN/MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_161.tif"
#> [2] "/tmp/RtmprleXzN/MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_177.tif"
if (length(outfiles_garda) > 0) {
plot(raster(outfiles_iseo[1]))
}
outfiles_iseo <- list.files(
file.path(tempdir(), "MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI"),
full.names = TRUE
)
outfiles_iseo
#> [1] "/tmp/RtmprleXzN/MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_161.tif"
#> [2] "/tmp/RtmprleXzN/MODIStsp/iseo_lake/VI_16Days_1Km_v61/NDVI/MOD13A2_NDVI_2016_177.tif"
if (length(outfiles_garda) > 0) {
plot(raster(outfiles_iseo[1]))
}
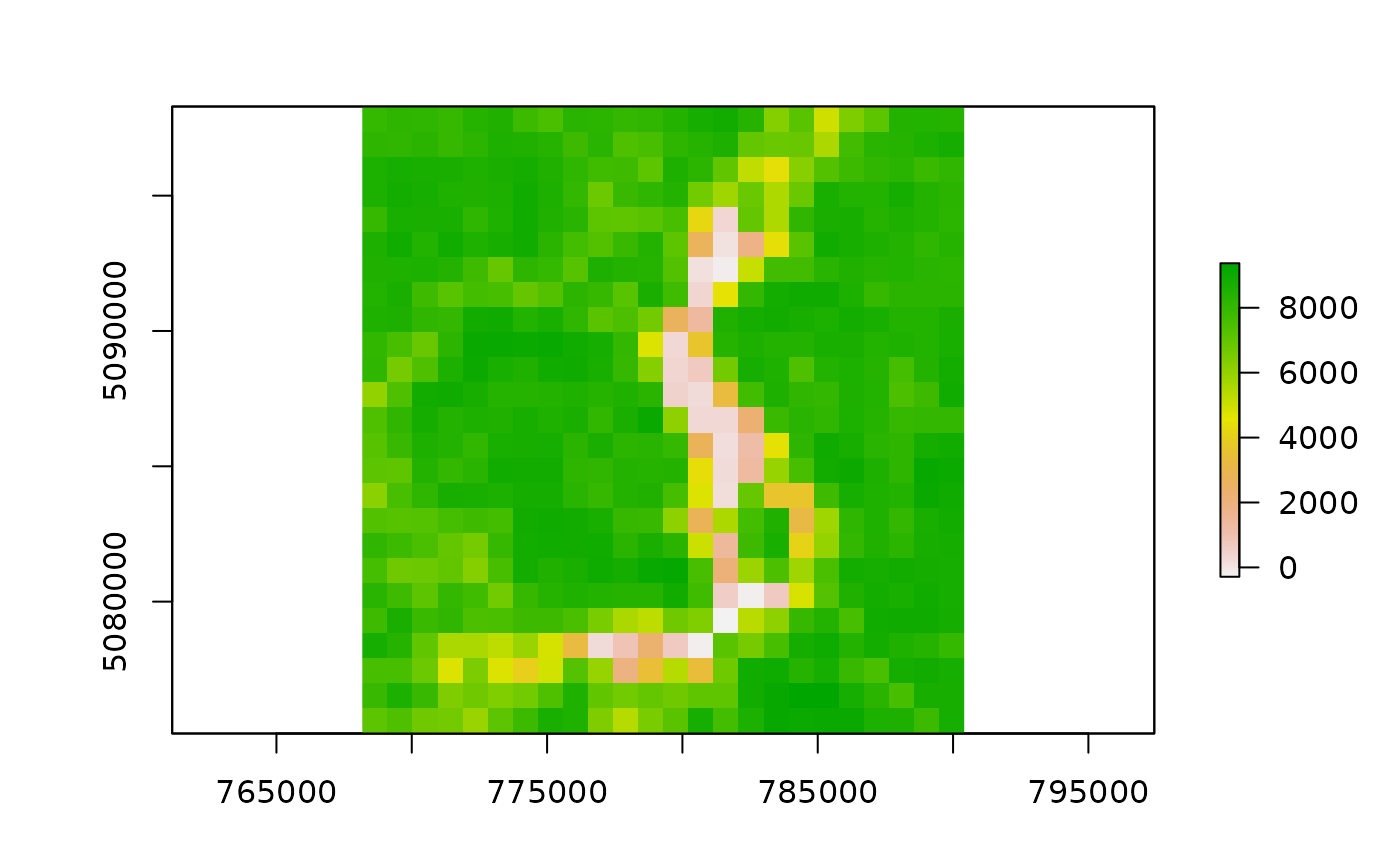 # See also https://docs.ropensci.org/MODIStsp/articles/noninteractive_execution.html
# }
# See also https://docs.ropensci.org/MODIStsp/articles/noninteractive_execution.html
# }
