Out-of-bag data
In random forests, each tree is grown with a bootstrapped version of the training set. Because bootstrap samples are selected with replacement, each bootstrapped training set contains about two-thirds of instances in the original training set. The ‘out-of-bag’ data are instances that are not in the bootstrapped training set.
Out-of-bag predictions and error
Each tree in the random forest can make predictions for its out-of-bag data, and the out-of-bag predictions can be aggregated to make an ensemble out-of-bag prediction. Since the out-of-bag data are not used to grow the tree, the accuracy of the ensemble out-of-bag predictions approximate the generalization error of the random forest. Out-of-bag prediction error plays a central role for some routines that estimate variable importance, e.g. negation importance.
We fit an oblique random survival forest and plot the distribution of the ensemble out-of-bag predictions.
fit <- orsf(data = pbc_orsf,
formula = Surv(time, status) ~ . - id,
oobag_pred_type = 'surv',
n_tree = 5,
oobag_pred_horizon = 2000)
hist(fit$pred_oobag,
main = 'Out-of-bag survival predictions at t=2,000')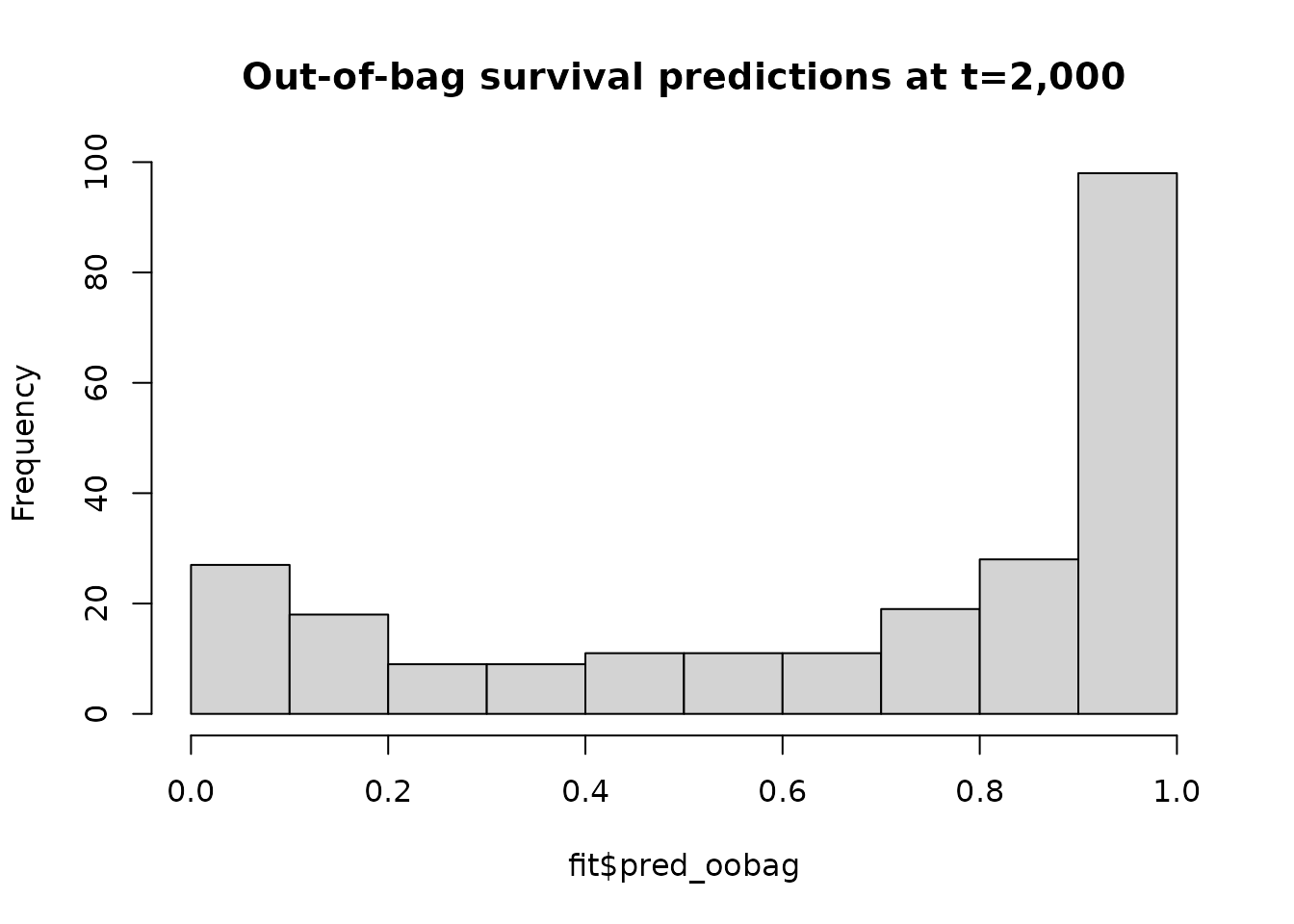
Next, let’s check the out-of-bag accuracy of fit:
# what function is used to evaluate out-of-bag predictions?
fit$eval_oobag$stat_type
#> [1] "Harrell's C-index"
# what is the output from this function?
fit$eval_oobag$stat_values
#> [,1]
#> [1,] 0.7419135The out-of-bag estimate of Harrell’s C-index (the default method to evaluate out-of-bag predictions) is 0.7419135.
Monitoring out-of-bag error
As each out-of-bag data set contains about one-third of the training
set, the out-of-bag error estimate usually converges to a stable value
as more trees are added to the forest. If you want to monitor the
convergence of out-of-bag error for your own oblique random survival
forest, you can set oobag_eval_every to compute out-of-bag
error at every oobag_eval_every tree. For example, let’s
compute out-of-bag error after fitting each tree in a forest of 50
trees:
fit <- orsf(data = pbc_orsf,
formula = Surv(time, status) ~ . - id,
n_tree = 20,
tree_seeds = 2,
oobag_pred_type = 'surv',
oobag_pred_horizon = 2000,
oobag_eval_every = 1)
plot(
x = seq(1, 20, by = 1),
y = fit$eval_oobag$stat_values,
main = 'Out-of-bag C-statistic computed after each new tree is grown.',
xlab = 'Number of trees grown',
ylab = fit$eval_oobag$stat_type
)
lines(x=seq(1, 20), y = fit$eval_oobag$stat_values)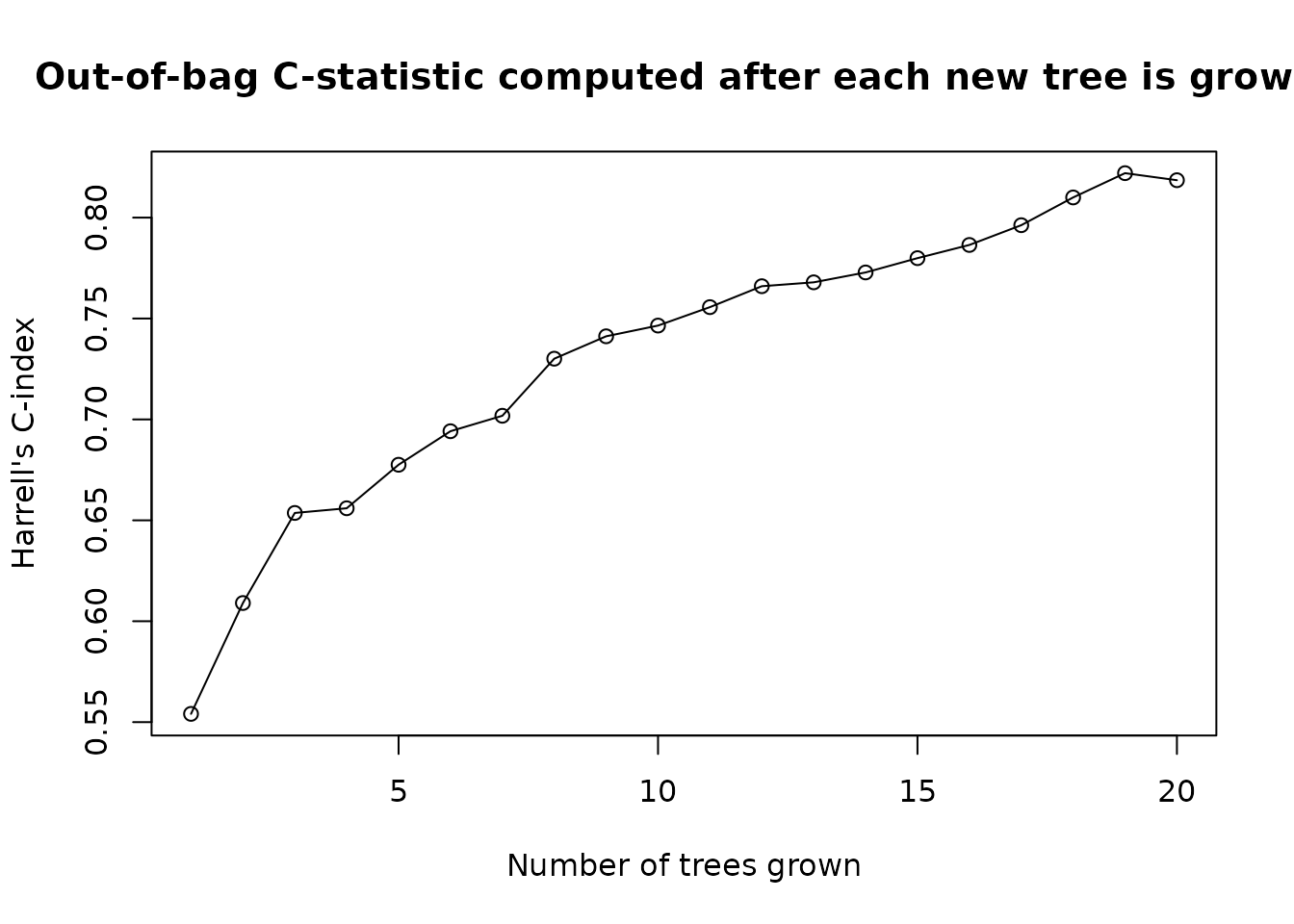
In general, at least 500 trees are recommended for a random forest fit. We’re just using 10 for illustration.
User-supplied out-of-bag evaluation functions
In some cases, you may want more control over how out-of-bag error is
estimated. For example, let’s use the Brier score from the
SurvMetrics package:
oobag_brier_surv <- function(y_mat, w_vec, s_vec){
# use if SurvMetrics is available
if(requireNamespace("SurvMetrics")){
return(
# output is numeric vector of length 1
as.numeric(
SurvMetrics::Brier(
object = Surv(time = y_mat[, 1], event = y_mat[, 2]),
pre_sp = s_vec,
# t_star in Brier() should match oob_pred_horizon in orsf()
t_star = 2000
)
)
)
}
# if not available, use a dummy version
mean( (y_mat[,2] - (1-s_vec))^2 )
}There are two ways to apply your own function to compute out-of-bag error. First, you can apply your function to the out-of-bag survival predictions that are stored in ‘aorsf’ objects, e.g:
oobag_brier_surv(y_mat = pbc_orsf[,c('time', 'status')],
s_vec = fit$pred_oobag)
#> Loading required namespace: SurvMetrics
#> [1] 0.11869Second, you can pass your function into orsf(), and it
will be used in place of Harrell’s C-statistic:
# instead of copy/pasting the modeling code and then modifying it,
# you can just use orsf_update.
fit_brier <- orsf_update(fit, oobag_fun = oobag_brier_surv)
plot(
x = seq(1, 20, by = 1),
y = fit_brier$eval_oobag$stat_values,
main = 'Out-of-bag error computed after each new tree is grown.',
sub = 'For the Brier score, lower values indicate more accurate predictions',
xlab = 'Number of trees grown',
ylab = "Brier score"
)
lines(x=seq(1, 20), y = fit_brier$eval_oobag$stat_values)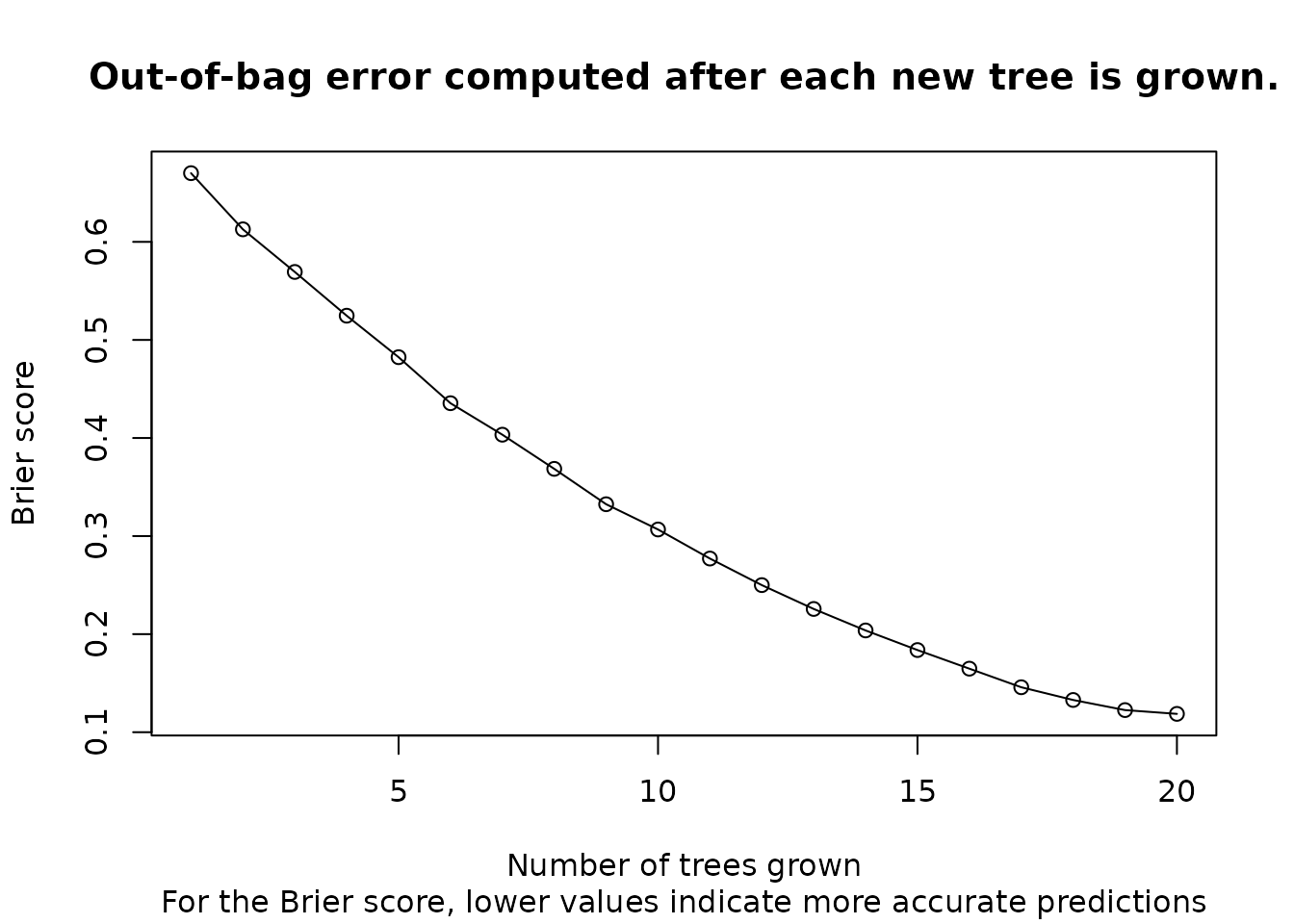
Specific instructions on user-supplied functions
if you use your own oobag_fun note the following:
oobag_funshould have three inputs:y_mat,w_vec, ands_vecFor survival trees,
y_matshould be a two column matrix with first column named ‘time’ and second named ‘status’. For classification trees,y_matshould be a matrix with number of columns = number of distinct classes in the outcome. For regression,y_matshould be a matrix with one column.s_vecis a numeric vector containing predictionsoobag_funshould return a numeric output of length 1
Notes
When evaluating out-of-bag error:
the
oobag_pred_horizoninput inorsf()determines the prediction horizon for out-of-bag predictions. The prediction horizon needs to be specified to evaluate prediction accuracy in some cases, such as the examples above. Be sure to check if that is the case when using your own functions, and if so, be sure thatoobag_pred_horizonmatches the prediction horizon used in your custom function.Some functions expect predicted risk (i.e., 1 - predicted survival), others expect predicted survival.
