
Basics of data import and cleaning in pathviewr
Vikram B. Baliga
2026-01-29
Source:vignettes/data-import-cleaning.Rmd
data-import-cleaning.RmdOverview
Raw movement data, including those from motion capture systems, may have a variety of issues. These raw data often contain noise or artifacts from the recording session, which may not be easily removed via the recording software itself. Data may not be organized as “tidy” key-value pairs (making plotting more difficult), the axes and overall orientation of the environment may not conform to a standard, and individual movement trajectories may be ill-defined.
pathviewr provides functions in R to deal with such
problems (i.e. “cleaning”). This vignette will cover the basics of how
to import raw data and how to clean data to prepare it for visualization
and/or statistical analyses.
What do movement data sets look like?
At minimum, movement data provide information on a subject or object’s position over time. These data are typically supplied in three dimensions (e.g. x, y, z), with position in each dimension sampled at a particular rate (e.g. 100 Hz). Different recording software may provide additional features, such as the ability to track multiple subjects simultaneously, information on subjects’ rotation, tracking of “rigid body” elements, or even the ability to apply Kalman filters.
A central goal of pathviewr is to take data from
different sources (so far: Motive and Flydra), re-organize them into a
common format that can be wrangled in R, clean them up a bit, and get
them ready for visualization and/or statistical analyses. We’ll first
cover what’s included in Motive and in Flydra data and how
pathviewr handles these. Should you have data from another
source, our as_viewr() function will allow you to bring it
into the pathviewr framework.
Data import via pathviewr
Data can be imported via one of three functions:
read_motive_csv()imports data from.csvfiles that have been exported from Optitrack’s Motive softwareread_flydra_mat()imports data from.matfiles that have been exported from Flydraas_viewr()can be used to handle data from other sources
We will showcase examples from each of these methods in this section.
Please feel free to reach out to the pathviewr authors
via our Github
Issues page should you have trouble with any of our data import
options. We are happy to work with you to design custom
read_ functions for file types we have not encountered
ourselves.
We’ll start by loading pathviewr and a few of the
packages in the tidyverse.
## If you do not already have pathviewr installed:
# install.packages("devtools")
# devtools::install_github("ropensci/pathviewr")
library(pathviewr)
library(ggplot2)
library(magrittr)Motive CSV files
.csv files exported from Motive can be imported via
read_motive_csv()
## Import the Motive example data included in
## the package
motive_data <-
read_motive_csv(
system.file("extdata", "pathviewr_motive_example_data.csv",
package = 'pathviewr')
)
## This produces a tibble
motive_data
#> # A tibble: 934 × 26
#> frame time_sec device02_rotation_x device02_rotation_y device02_rotation_z
#> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 72210 722. 0.135 -0.977 -0.112
#> 2 72211 722. 0.0819 -0.978 -0.0991
#> 3 72212 722. 0.211 -0.973 -0.0939
#> 4 72213 722. 0.196 -0.972 -0.128
#> 5 72214 722. 0.131 -0.975 -0.121
#> 6 72215 722. 0.0935 -0.975 -0.105
#> 7 72216 722. 0.180 -0.975 -0.106
#> 8 72217 722. 0.164 -0.972 -0.149
#> 9 72218 722. 0.120 -0.973 -0.149
#> 10 72219 722. 0.155 -0.970 -0.120
#> # ℹ 924 more rows
#> # ℹ 21 more variables: device02_rotation_w <dbl>, device02_position_x <dbl>,
#> # device02_position_y <dbl>, device02_position_z <dbl>,
#> # device02_mean_marker_error <dbl>, device03_rotation_x <dbl>,
#> # device03_rotation_y <dbl>, device03_rotation_z <dbl>,
#> # device03_rotation_w <dbl>, device03_position_x <dbl>,
#> # device03_position_y <dbl>, device03_position_z <dbl>, …A key thing to note is that these data, as stored in Motive CSVs, are
not “tidy”. Each frame occupies one row, but what that also means is
that the rotation and position values for the various subjects take up
24 columns! This format not only makes plotting data more difficult in
base R, ggplot2, and rgl, but also makes other
aspects of data wrangling more difficult. In a later step, we will
‘gather’ these data into key-value pairs so that e.g. all length-wise
position values are in one column, all width-wise are in
another…etc.
Metadata are stored as attributes. We won’t go through all of these, but here are a couple important ones.
## E.g. to see the header of the original file:
attr(motive_data, "header")
#> metadata value
#> 1 Format Version 1.23
#> 2 Take Name sept-18_mixed-group_16-30
#> 3 Take Notes
#> 4 Capture Frame Rate 100.000000
#> 5 Export Frame Rate 100.000000
#> 6 Capture Start Time 2019-09-18 04.30.02.695 PM
#> 7 Total Frames in Take 190951
#> 8 Total Exported Frames 190951
#> 9 Rotation Type Quaternion
#> 10 Length Units Meters
#> 11 Coordinate Space Global
## Names of all marked objects:
attr(motive_data, "subject_names_simple")
#> [1] "device02" "device03" "device05"
## Types of data included
attr(motive_data, "data_types_simple")
#> [1] "Rigid Body"
## Frame rate
attr(motive_data, "frame_rate")
#> [1] 100Storing such metatdata in the attributes is a key feature of
pathviewr. These metadata may not be as immediately as
important as the time series of position or rotation, but they can
provide important experimental information such as the date & time
of capture and the units of the position data (here, meters).
Flydra Matlab files
.mat files exported from Flydra can be imported via
read_flydra_mat().
Note that you must supply a subject_name for Flydra
data, as subject names are not embedded in the .mat files.
Only one name can be added and it will be used throughout the resultant
tibble.
## Import the Flydra example data included in
## the package
flydra_data <-
read_flydra_mat(
system.file("extdata",
"pathviewr_flydra_example_data.mat",
package = 'pathviewr'),
subject_name = "birdie_wooster"
)
## Similarly, this produces a tibble with important
## metadata as attributes
flydra_data
#> # A tibble: 7,744 × 10
#> frame time_sec subject position_length position_width position_height
#> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 746 0 birdie_wooster 0.869 -0.00417 1.31
#> 2 747 0.01 birdie_wooster 0.864 -0.00614 1.31
#> 3 748 0.02 birdie_wooster 0.863 -0.00695 1.31
#> 4 749 0.03 birdie_wooster 0.862 -0.00672 1.31
#> 5 750 0.04 birdie_wooster 0.862 -0.00644 1.31
#> 6 751 0.05 birdie_wooster 0.862 -0.00619 1.31
#> 7 752 0.06 birdie_wooster 0.863 -0.00667 1.31
#> 8 753 0.07 birdie_wooster 0.864 -0.00712 1.31
#> 9 754 0.08 birdie_wooster 0.865 -0.00727 1.31
#> 10 755 0.09 birdie_wooster 0.865 -0.00760 1.31
#> # ℹ 7,734 more rows
#> # ℹ 4 more variables: velocity <dbl>, length_inst_vel <dbl>,
#> # width_inst_vel <dbl>, height_inst_vel <dbl>
attr(flydra_data, "frame_rate")
#> [1] 100Note that unlike the example Motive data, the Flydra data are already organized into key-value pairs. Because rotation is not captured by Flydra, such data are also not included.
Data from other sources
Data from another format can be converted to a viewr
object via pathviewr::as_viewr(). Although this function
does not handle data import per se, it allows data that you may already
have imported into R as a tibble or data.frame
to then be reformatted for use with pathviewr
functions.
We’ll run through a quick example with simulated data:
## Create a dummy data frame with simulated (nonsense) data
df <-
data.frame(
frame = seq(1, 100, by = 1),
time_sec = seq(0, by = 0.01, length.out = 100),
subject = "birdie_sanders",
z = rnorm(100),
x = rnorm(100),
y = rnorm(100)
)
## Use as_viewr() to convert it into a viewr object
test <-
as_viewr(
df,
frame_rate = 100,
frame_col = 1,
time_col = 2,
subject_col = 3,
position_length_col = 5,
position_width_col = 6,
position_height_col = 4
)
## Some metadata are stored as attributes
attr(test, "frame_rate")
#> [1] 100We also welcome you to request custom data import functions,
especially if as_viewr() does not fit your needs. We are
interested in expanding our data import functions to accommodate
additional file types. Please feel free to file a
request for additional import functions via our Github Issues
page.
Data cleaning
As noted above, raw data often suffer the following:
- contain noise or artifacts from the recording session
- not organized as “tidy” key-value pairs
- axes and overall orientation of the environment may not conform to a
standard
- individual movement trajectories may be ill-defined
Several functions to clean and wrangle data are available, and we have a suggested pipeline for how these steps should be handled. The rest of this vignette will cover these steps.
All of the steps in the suggested pipeline are also covered by two
all-in-one functions: clean_viewr() and
import_and_clean_viewr(). See the section at the very end
of this vignette for details.
And speaking of pipes, all functions in pathviewr are
pipe-friendly. We will detail each step separately, but each of the
subsequent steps may be piped, e.g.
data %>% relabel_viewr_axes() %>% gather_tunnel_data()
etc etc
Relabeling axes, gathering data columns, and trimming outliers
Axis labels (x, y, z) may be applied in arbitrary ways by software. A user might intuitively think the z axis represents height, but the original software may label it as the y axis instead.
relabel_viewr_axes provides a means to relabel axes with
“tunnel_length”, “tunnel_width”, and “tunnel_height”. These axis
labels will be expected by subsequent functions, so skipping this step
is ill-advised.
Typically, axes from Motive data will need to be relabled, but axes in data imported from Flydra will not.
motive_relabeled <-
motive_data %>%
relabel_viewr_axes(
tunnel_length = "_z",
tunnel_width = "_x",
tunnel_height = "_y",
real = "_w"
)
names(motive_relabeled)
#> [1] "frame" "time_sec"
#> [3] "device02_rotation_width" "device02_rotation_height"
#> [5] "device02_rotation_length" "device02_rotation_real"
#> [7] "device02_position_width" "device02_position_height"
#> [9] "device02_position_length" "device02_mean_marker_error"
#> [11] "device03_rotation_width" "device03_rotation_height"
#> [13] "device03_rotation_length" "device03_rotation_real"
#> [15] "device03_position_width" "device03_position_height"
#> [17] "device03_position_length" "device03_mean_marker_error"
#> [19] "device05_rotation_width" "device05_rotation_height"
#> [21] "device05_rotation_length" "device05_rotation_real"
#> [23] "device05_position_width" "device05_position_height"
#> [25] "device05_position_length" "device05_mean_marker_error"Akin to the behavior of dplyr::gather(),
gather_tunnel_data() will take all data from a given
session and organize it so that all data of a given type are within one
column, i.e. all position lengths are in position_length,
as opposed to separate length columns for each rigid body. These
column names will be expected by subsequent functions, so skipping this
step is also ill-advised if you are using data from Motive.
Should you have data from Flydra, this step should be skipable.
Use trim_tunnel_outliers() to remove extreme artifacts
and other outlier data. What this function does is create a (virtual)
boundary box according to user-specification, and any data outside that
boundary are removed. For example, if you know your arena measures 10m x
10m x 10m and your data were calibrated to range from 0-10m in each
dimension, you can be reasonably sure that extreme values such as 45m on
a given axis are bogus. This step is entirely optional, and should only
be used when the user is confident that data outside certain ranges are
artifacts or other bugs. Data outside these ranges are then filtered
out. Best to plot data beforehand and check!!
## First gather and show the new column names
motive_gathered <-
motive_relabeled %>%
gather_tunnel_data()
names(motive_gathered)
#> [1] "frame" "time_sec" "subject"
#> [4] "position_length" "position_width" "position_height"
#> [7] "rotation_length" "rotation_width" "rotation_height"
#> [10] "rotation_real" "mean_marker_error"
## Now trim, using ranges we know to safely include data
## and exclude artifacts. Anything outside these ranges
## will be removed.
motive_trimmed <-
motive_gathered %>%
trim_tunnel_outliers(
lengths_min = 0,
lengths_max = 3,
widths_min = -0.4,
widths_max = 0.8,
heights_min = -0.2,
heights_max = 0.5
)Standardization of tunnel position and coordinates
The coordinate system of the tunnel itself may require adjustment or
standardization. For example, data collected across different days may
show slight differences in coordinate systems if calibration equipment
was not used in identical ways. Moreover, the user may want to redefine
how the coordinate system itself is defined (i.e. change the location of
(0, 0, 0) to another place within the tunnel.
Note that having (0, 0, 0) set to the center of the
region of interest (covered in the next section of this vignette) is
required for all subsequent pathviewr functions to
work.
pathviewr offers three main choices for such
standardization:
redefine_tunnel_center(): Sets the location of 0 on any or all axes to a new location. See the Help page for this function to see the four different methods by which a user can specify this. No rotation of the tunnel is performed. This function can be used on both Motive and Flydra data.standardize_tunnel(): Use specified landmarks (subjectswithin theviewrobject) to rotate and translate the location of a tunnel, setting(0, 0, 0)to the center of the tunnel (centering). For example, in an avian flight tunnel, perches may be set up on opposite ends of the tunnel and rigid body markers may be set to them. The positions of these perches can be used as landmarks to standardize tunnel position. Note that this is typically not possible for Flydra data, since Flydra data will be imported with only onesubject.rotate_tunnel: Rotate and center a tunnel based on user-defined coordinates (i.e. similar tostandardize_tunnel()but for cases where specified landmarks are not in the data). This function can be used on both Motive and Flydra data.
Two quick examples will follow, using our Motive and Flydra data:
## Rotate and center the motive data set:
motive_rotated <-
motive_trimmed %>%
rotate_tunnel(
perch1_len_min = -0.06,
perch1_len_max = 0.06,
perch2_len_min = 2.48,
perch2_len_max = 2.6,
perch1_wid_min = 0.09,
perch1_wid_max = 0.31,
perch2_wid_min = 0.13,
perch2_wid_max = 0.35
)In the above, virtual perches are defined by the user using the
arguments shown. The center of each perch is then found and then the
locations of the two perch centers are then used to 1) set
(0, 0, 0) to the point that is equidistant from the perches
(i.e. the middle of the tunnel) and 2) rotate the tunnel about the
height axis so that both perch width coordinates are at 0. This may be
easier to understand through plotting:
## Quick (base-R) plot of the original data
plot(motive_trimmed$position_length,
motive_trimmed$position_width,
asp = 1)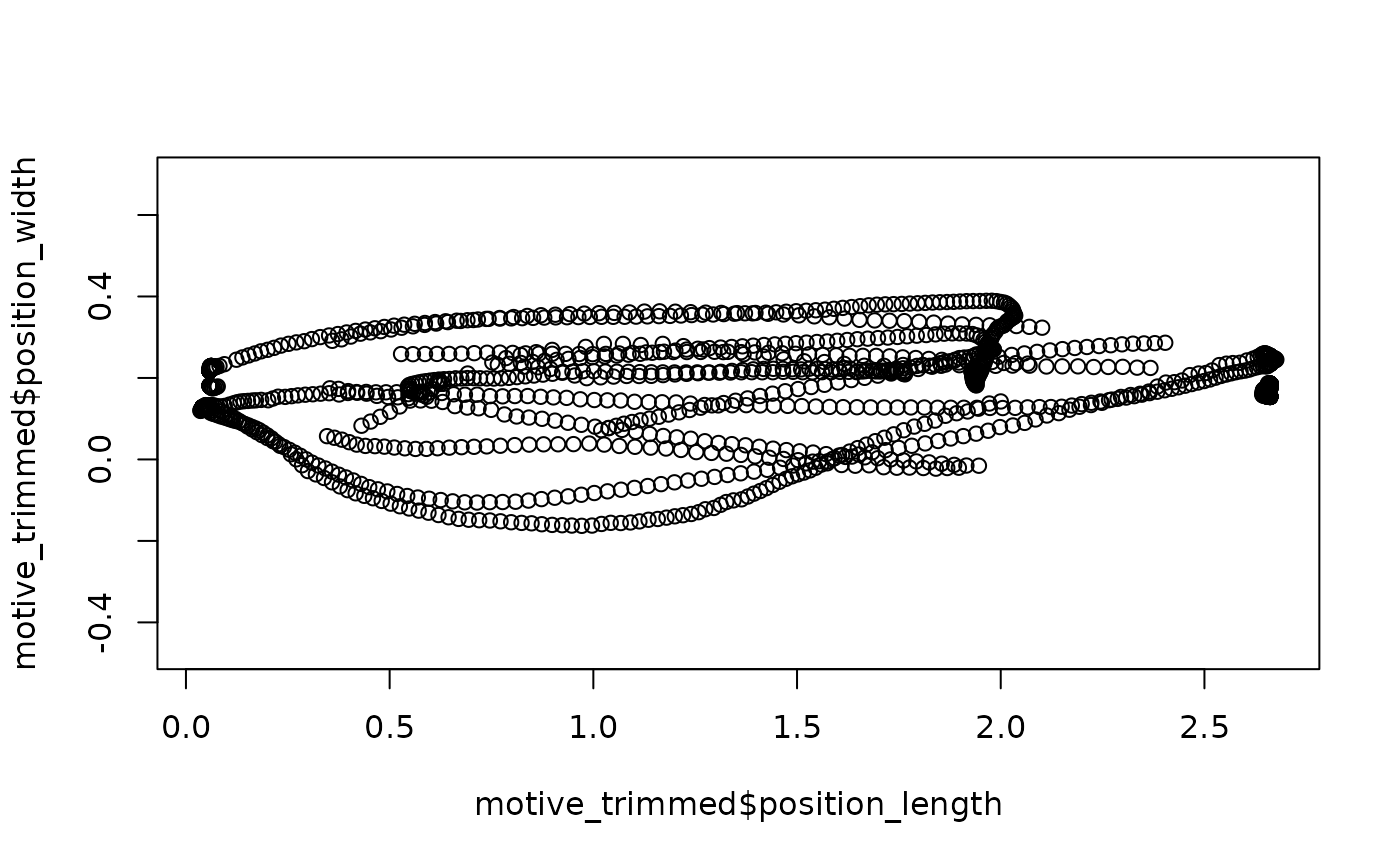
## Quick (base-R) plot of the rotated data
plot(motive_rotated$position_length,
motive_rotated$position_width,
asp = 1)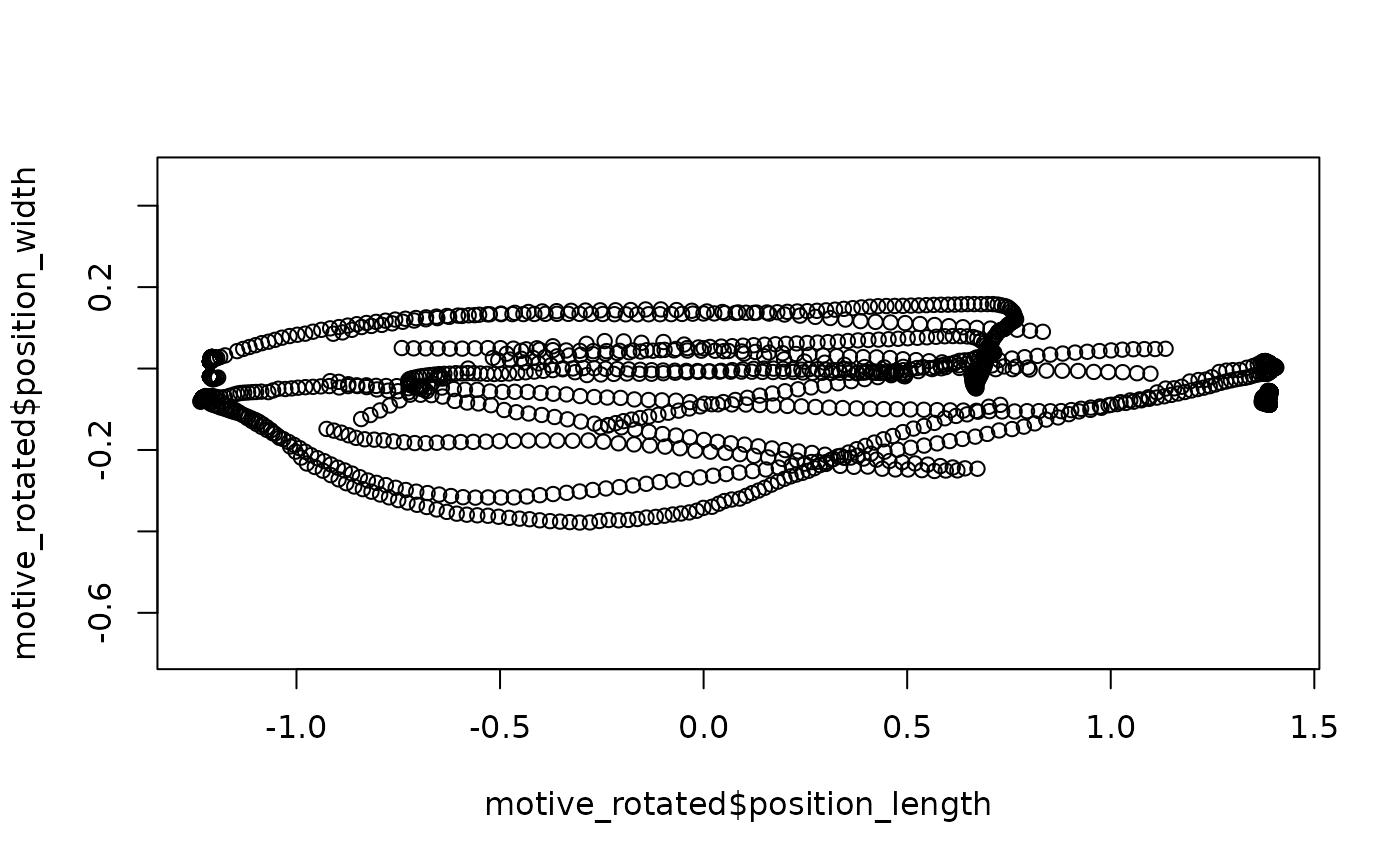
Differences due to rotation may be extremely subtle, but the
redefining of (0, 0, 0) to the middle of the tunnel should
be clear from contrasting the axes of the plots.
Flydra data typically do not need to be rotated, so we will instead
use redefine_tunnel_center() to adjust the location of
(0, 0, 0):
## Re-center the Flydra data set:
flydra_centered <-
flydra_data %>%
redefine_tunnel_center(length_method = "middle",
height_method = "user-defined",
height_zero = 1.44)Here, we are using length_method = "middle" to use the
middle of the range of “length” data to set length = 0 (a translation),
making no change to the width axis, and then specifying that height = 0
should be equal to the value 1.44 from the original data
(another translation). Again, plotting may help; note that this time,
we’ll plot length x height (instead of width):
## Quick (base-R) plot of the original data
plot(flydra_data$position_length,
flydra_data$position_height,
asp = 1)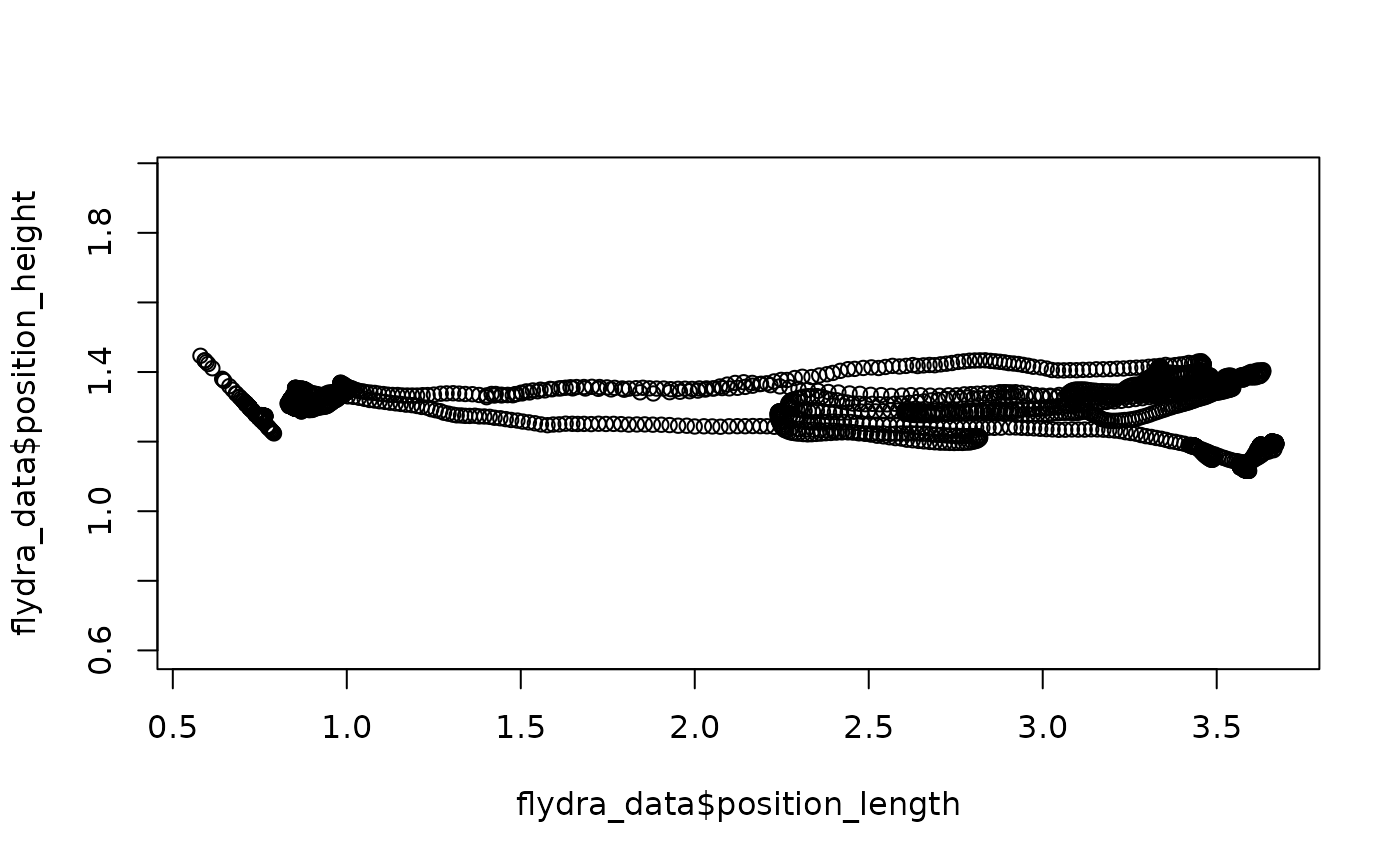
## Quick (base-R) plot of the redefined data
plot(flydra_centered$position_length,
flydra_centered$position_height,
asp = 1)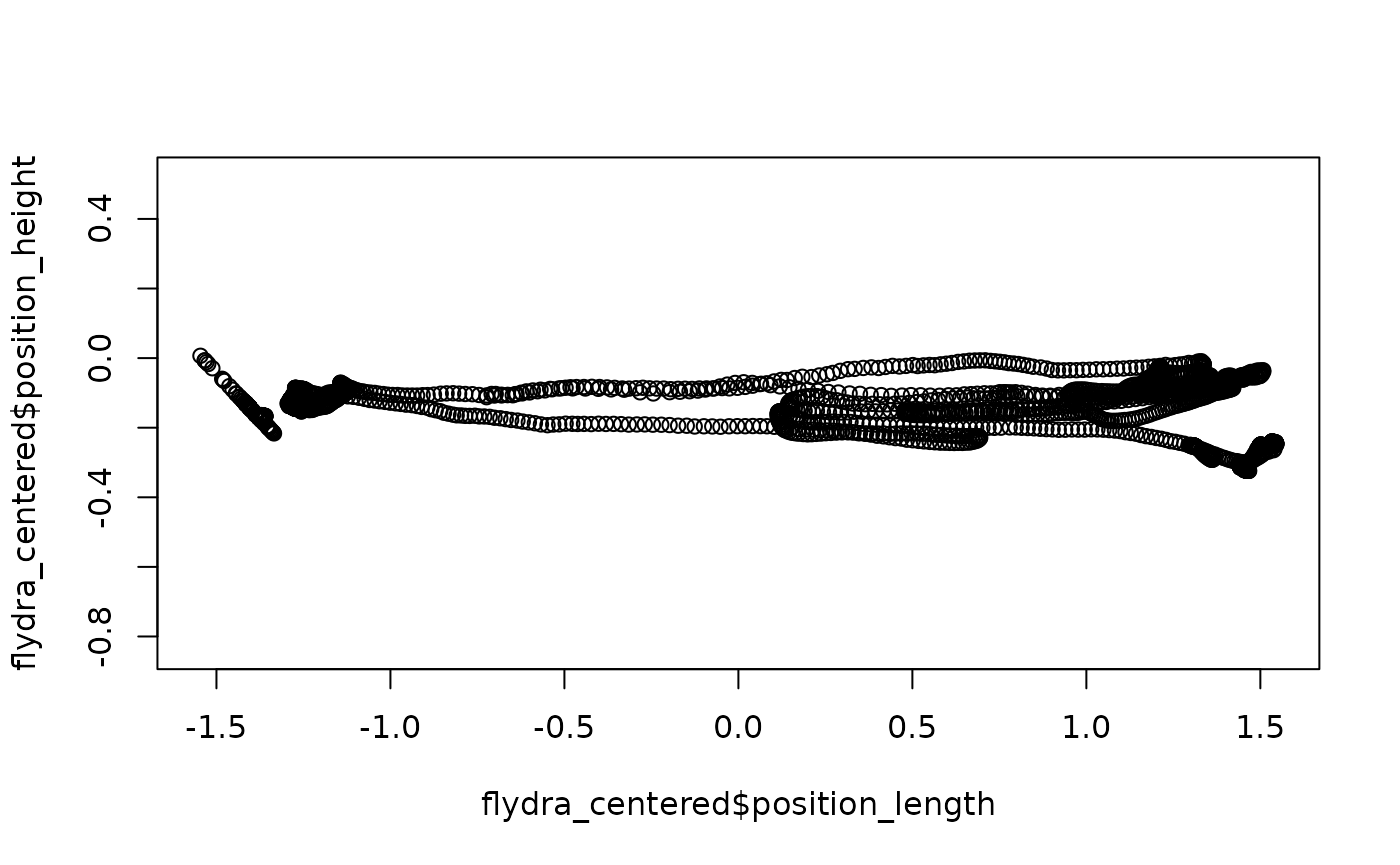
Selecting a region of interest
This required step has benefits that are twofold: 1) treatment effects on animal movement may only manifest over certain regions of the tunnel, and 2) focusing on a subset of the data makes it easier to define explicit trajectories and run computations faster.
The region of interest is defined via the function
select_x_percent(). Once tunnel coordinates have been
standardized (via one of the function in the previous section),
select_x_percent() then selects the middle x
percent (along the length axis) of the tunnel as the region of interest.
For example, selecting 50 percent would start from the center of the
tunnel and move 25% of the tunnel along positive length and 25% along
negative length values to then select the middle 50% of the tunnel.
Quick examples:
## Motive data: select the middle 50% of the tunnel as the region of interest
motive_selected <-
motive_rotated %>% select_x_percent(50)
## Quick plot:
plot(motive_selected$position_length,
motive_selected$position_width,
asp = 1)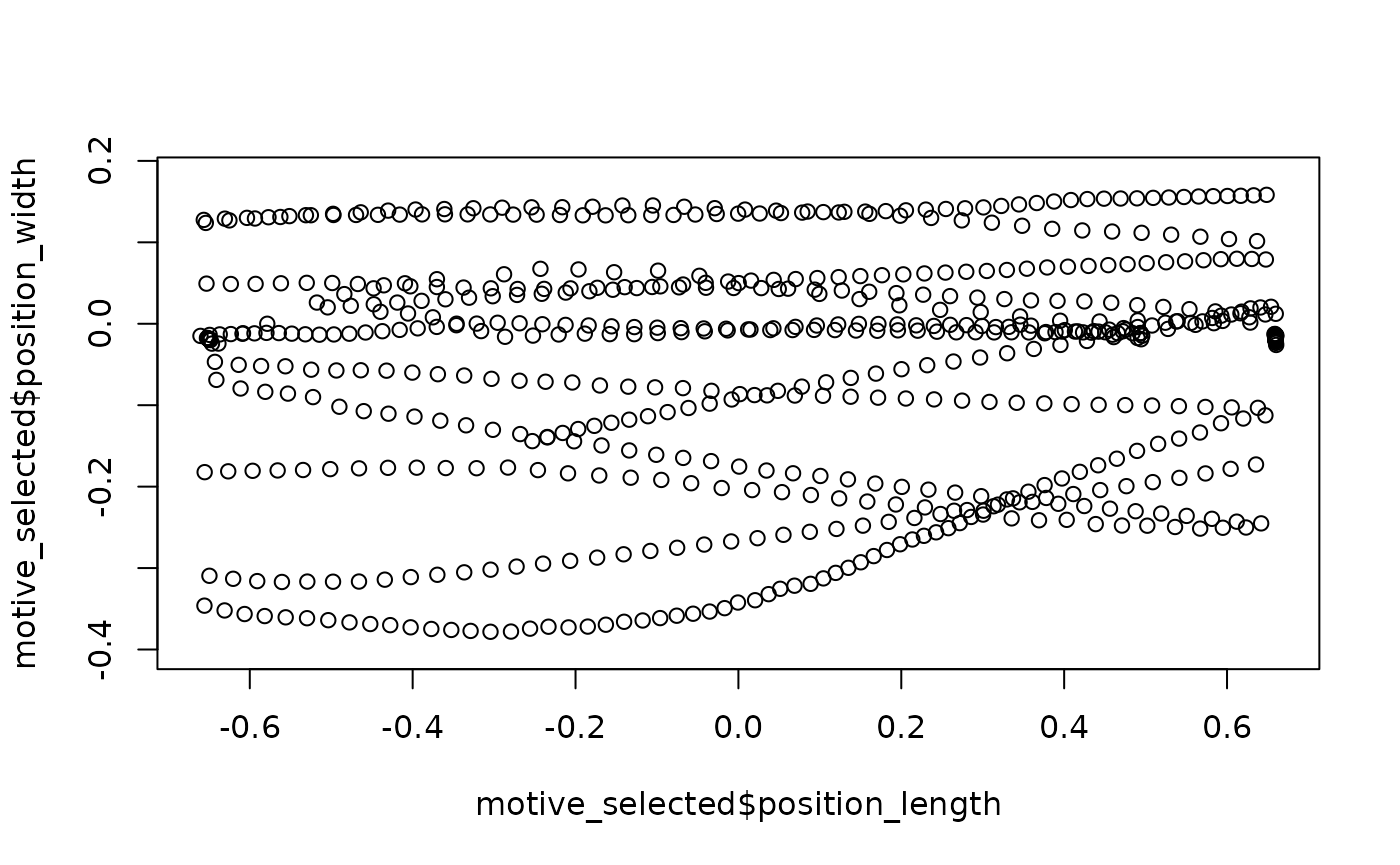
## Flydra data:
flydra_selected <-
flydra_centered %>% select_x_percent(50)
## Quick plot:
plot(flydra_selected$position_length,
flydra_selected$position_width,
asp = 1)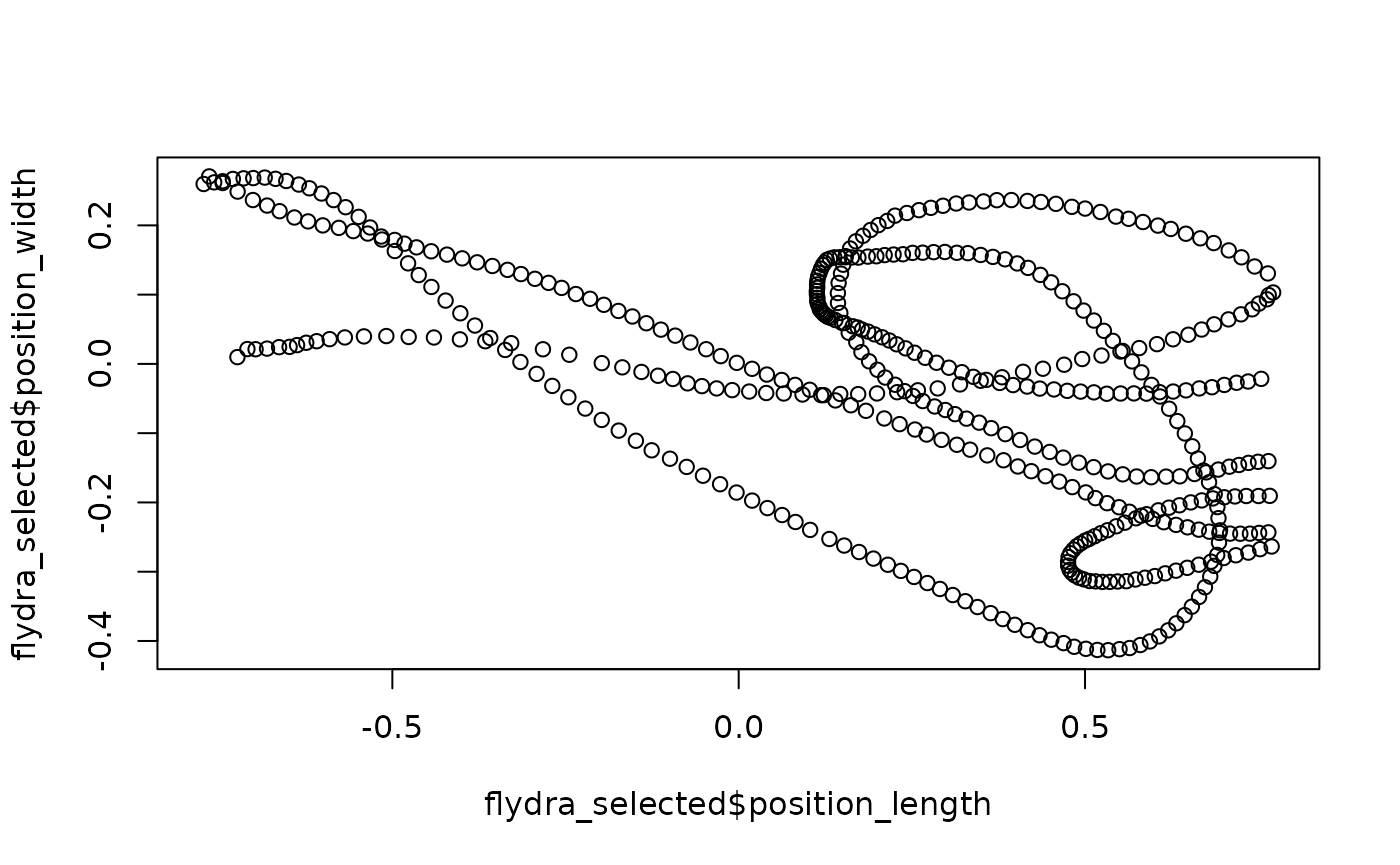
Isolating each trajectory
The pathviewr standard for defining a trajectory is:
continuous movement from one side of the tunnel to the other over the
span of the region of interest. Note that this definition does not
strictly require linear movement from one end to the other; an animal
could make several loops inside the region of interest within a given
trajectory.
Isolating trajectories is handled via the
separate_trajectories() function in pathviewr.
Note that a region of interest must be selected beforehand via
select_x_percent().
Because cameras may occasionally drop frames, we allow the user to
permit some relaxation of how stringent the “continuous movement”
criterion is. This is handled via the max_frame_gap
argument within separate_trajectories(). For more details,
please see the
vignette Managing frame gaps with pathviewr.
In our Motive example, we’ll use the automated feature built into the
function to guesstimate the best max_frame_gap allowed.
When frame gaps larger than max_frame_gap are encountered,
the function will force the defining of a new trajectory. But if frame
gaps smaller than max_frame_gap are encountered, keeping
observations within the same trajectory is permitted.
In the Flydra example, we’ll simply set max_frame_gap to
1 so that no frame gaps are allowed (movement must be
continuous with no dropped frames).
motive_labeled <-
motive_selected %>%
separate_trajectories(max_frame_gap = "autodetect")
#> autodetect is an experimental feature -- please report issues.
flydra_labeled <-
flydra_selected %>%
separate_trajectories(max_frame_gap = 1)Retain only complete trajectories
Now that trajectories have been isolated and labeled, the final cleaning step is to retain only the trajectories that completely span from one end of the region of interest to the other.
This final step is handled via pathviewr’s
get_full_trajectories().
There is a built-in “fuzziness” feature: because trajectories may not
have observations exactly at the beginning or the end of the region of
interest, it may be necessary to allow trajectories to be slightly
shorter than the range of the selected region of interest. The
span parameter of this function handles this. By supplying
a numeric proportion from 0 to 1, a user may
allow trajectories to span that proportion of the selected region. For
example, setting span = 0.95 will keep all trajectories
that span 95% of the length of the selected region of interest. Setting
span = 1 (not recommended) will strictly keep trajectories
that start and end at the exact cut-offs of the
selected region of interest.
For these reasons, spans of 0.99 to
0.95 are generally recommended. The best choice ultimately
depends on your capture frame rate as well as your own judgment. Should
you desire to set it lower (which you can), you may instead consider
using a smaller region of interest (i.e. set the
desired_percent parameter in
select_x_percent() to be lower).
## Motive
motive_full <-
motive_labeled %>%
get_full_trajectories(span = 0.95)
plot(motive_full$position_length,
motive_full$position_width,
asp = 1, col = as.factor(motive_full$file_sub_traj))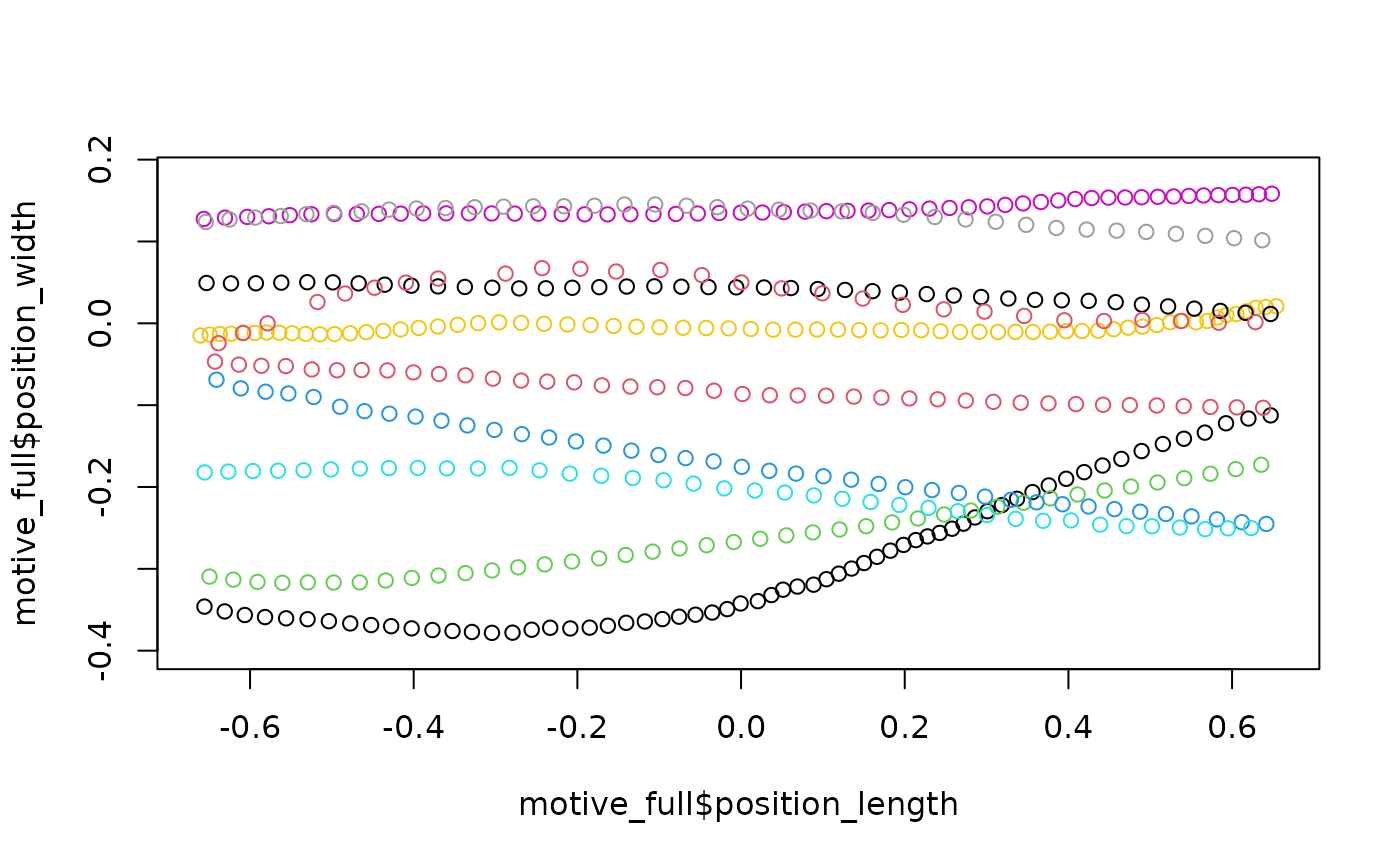
## Flydra
flydra_full <-
flydra_labeled %>%
get_full_trajectories(span = 0.95)
plot(flydra_full$position_length,
flydra_full$position_width,
asp = 1, col = as.factor(flydra_full$file_sub_traj))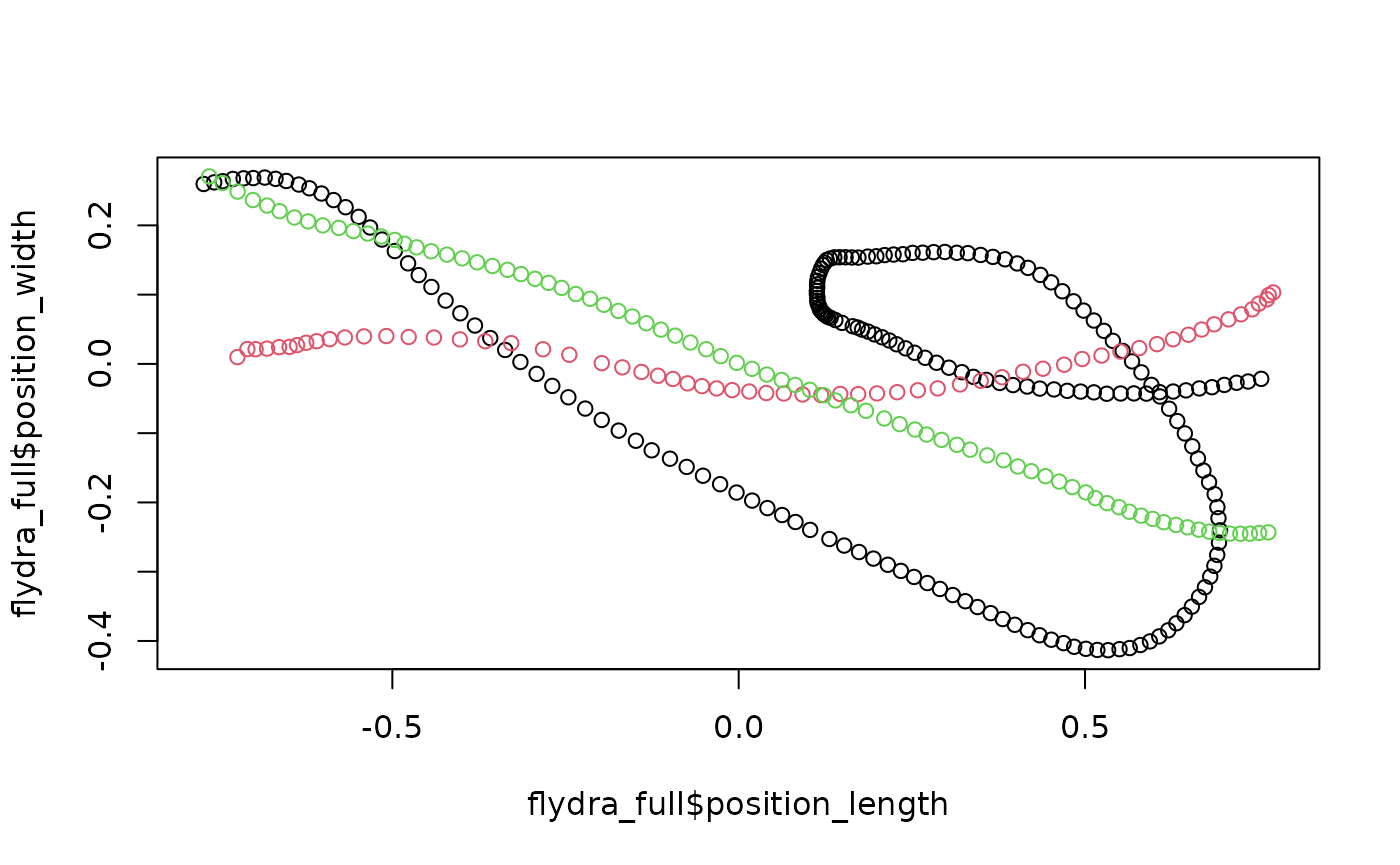
All-in-one cleaning functions
All of the above steps can also be done by using
pathviewr’s designated all-in-one functions.
import_and_clean_viewr() imports raw data and allows the
user to run through all of the cleaning steps previously covered in this
vignette. clean_viewr() does the same on any object already
imported into the R environment (i.e. it skips data import).
For both of these functions, all of the cleaning steps are set to
TRUE by default, but may be turned off by using
FALSE. Argument names correspond to standalone functions in
pathviewr, and if the user wants to use non-default values
for corresponding arguments, they should also be supplied for any steps
that are set to TRUE.
For example, if the user keeps select_x_percent = TRUE
as an argument in clean_viewr(), the
select_x_percent() function is run internally. This means
that should the user desire to select a region of interest that does not
match the default value of 33 percent, an additional argument should be
supplied to clean_viewr() as if it were being supplied to
select_x_percent() itself, e.g.:
desired_percent = 50.
All additional arguments should be written out fully and explicitly.
Here’s an example using what we previously covered:
motive_allinone <-
motive_data %>%
clean_viewr(
relabel_viewr_axes = TRUE,
gather_tunnel_data = TRUE,
trim_tunnel_outliers = TRUE,
standardization_option = "rotate_tunnel",
select_x_percent = TRUE,
desired_percent = 50,
rename_viewr_characters = FALSE,
separate_trajectories = TRUE,
max_frame_gap = "autodetect",
get_full_trajectories = TRUE,
span = 0.95
)
#> autodetect is an experimental feature -- please report issues.
## Quick plot
plot(motive_allinone$position_length,
motive_allinone$position_width,
asp = 1, col = as.factor(motive_allinone$file_sub_traj))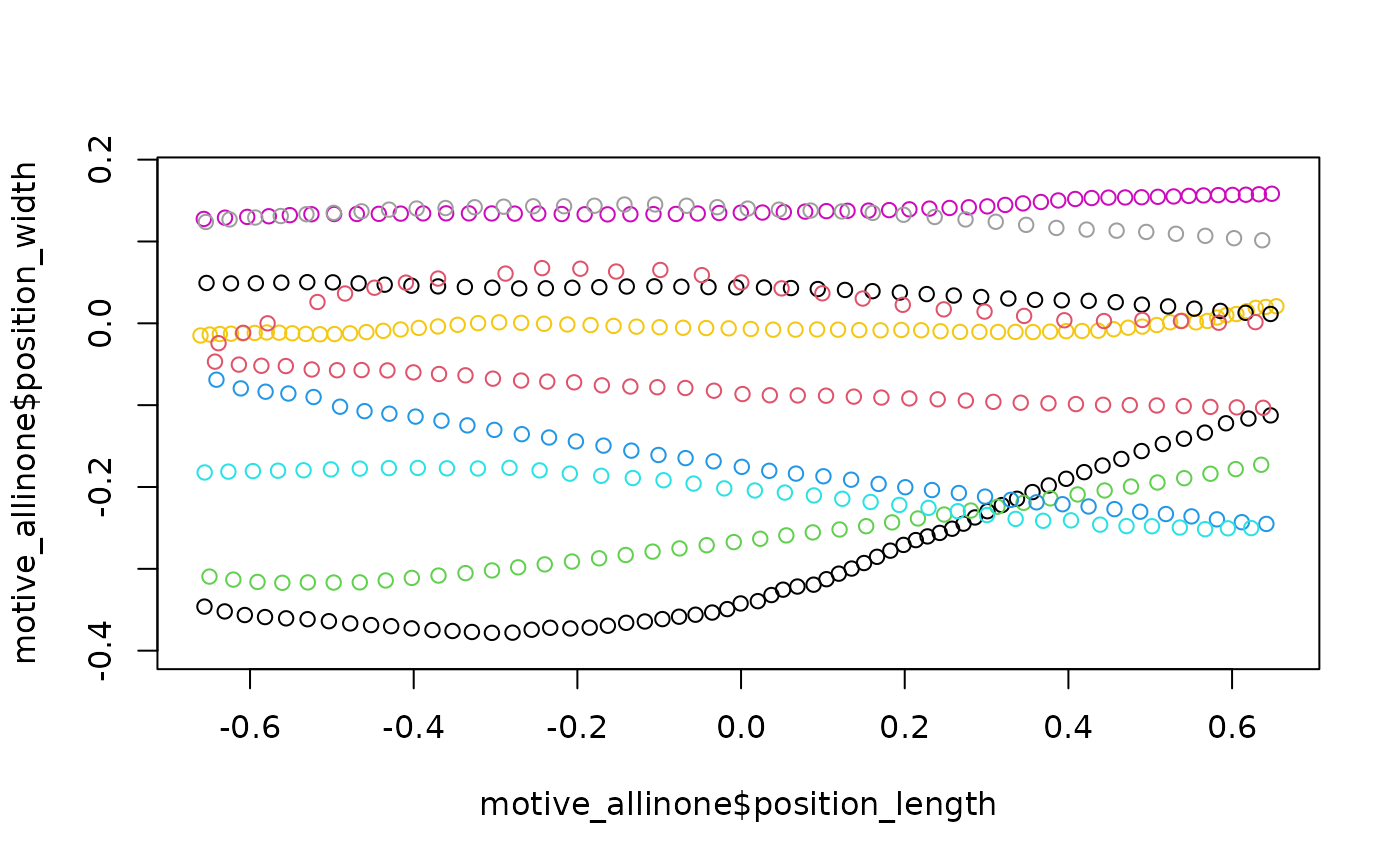
That’s all!
🐢