
Applying taxlist to species lists on diversity records
Miguel Alvarez
2026-03-05
Source:vignettes/taxlist-intro.Rmd
taxlist-intro.Rmd1. Getting started
The package taxlist aims to implement an object class
and functions (methods) for handling taxonomic data in
R. The homonymous object class taxlist can
be further linked to biodiversity records (e.g. for observations in
vegetation plots).
The taxlist package is developed on the repository
GitHub (https://github.com/ropensci/taxlist)
and can be installed in your R-session using the package
devtools:
Since this package is already available in the Comprehensive R
Archive Network (CRAN), it is also possible to install it using the
function install.packages:
install.packages("taxlist", dependencies = TRUE)Of course, you have to load taxlist into your
R-session.
For accessing to this vignette, use following command:
vignette("taxlist-intro")2. Extracting a species list from a vegetation table
2.1 Example data
One of the main tasks of taxlist is to structure
taxonomic information for a further linkage to biodiversity records.
This structure have to be on the one side consistent with taxonomic
issues (e.g. synonyms, hierarchies, etc.), on the other side have to be
flexible for containing different depth of information availability
(from plain species lists to hierarchical structures).
In this guide, we will work with a species list from phytosociological relevés collected at the borderline between the Democratic Republic of the Congo and Rwanda (Mullenders 1953 Vegetatio 4(2): 73–83).
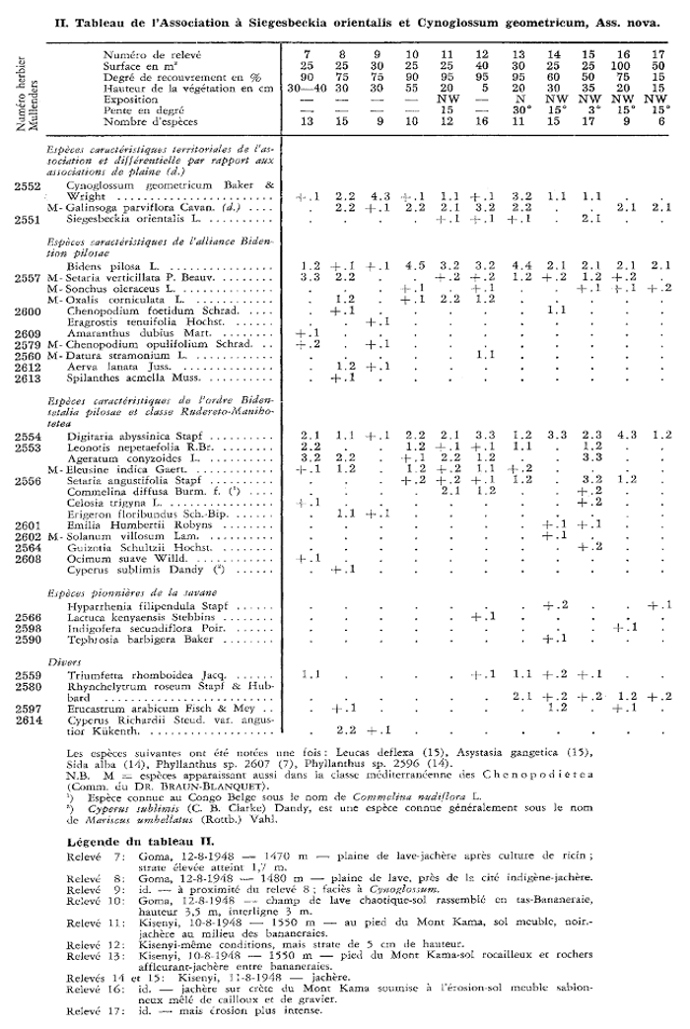
The digitized data can be loaded by following command:
load(file.path(path.package("taxlist"), "Cross.rda"))The data is formatted as data.frame in
R, including the names of the species in the first
column:
head(Cross[, 1:8])## TaxonName 3094 3093 3092 3095 3096 3097 3098
## 1 Eragrostis tenuifolia + <NA> <NA> <NA> <NA> <NA> <NA>
## 2 Cyperus sublimis <NA> + <NA> <NA> <NA> <NA> <NA>
## 3 Digitaria abyssinica + 1 2 2 2 3 1
## 4 Hyparrhenia filipendula <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 5 Erigeron floribundus + 1 <NA> <NA> <NA> <NA> <NA>
## 6 Aerva lanata + 1 <NA> <NA> <NA> <NA> <NA>2.2 From plain list to taxlist
As already mentioned, the first column in the cross table contains
the names of the species occurring in the observed plots. Thus, we can
use this character vector to construct a taxlist object.
This can be achieved through the function df2taxlist().
sp_list <- Cross[, "TaxonName"]
sp_list <- df2taxlist(x = sp_list)## Missing column 'TaxonConceptID' in 'x'. All names will be considered as accepted names.
summary(sp_list)## object size: 9 Kb
## validation of 'taxlist' object: TRUE
##
## number of taxon usage names: 35
## number of taxon concepts: 35
## trait entries: 0
## number of trait variables: 0
## taxon views: 0Note that the function summary provides a quick overview
in the content of the resulting object. This function can be also
applied to a specific taxon:
summary(object = sp_list, ConceptID = "Erigeron floribundus")## ------------------------------
## concept ID: 5
## view ID: none
## level: none
## parent: none
##
## # accepted name:
## 5 Erigeron floribundus NA
## ------------------------------3. Built-in data set
3.1 Easplist
The installation of taxlist includes the data
Easplist, which is formatted as a taxlist
object. This data is a subset of the species list used by the database
SWEA-Dataveg (GIVD ID
AF-006):
data(Easplist)
Easplist## object size: 761.4 Kb
## validation of 'taxlist' object: TRUE
##
## number of taxon usage names: 5393
## number of taxon concepts: 3887
## trait entries: 311
## number of trait variables: 1
## taxon views: 3
##
## concepts with parents: 3698
## concepts with children: 1343
##
## concepts with rank information: 3887
## concepts without rank information: 0
##
## family: 186
## genus: 1011
## complex: 1
## species: 2521
## subspecies: 71
## variety: 95
## form: 23.2 Access to slots
The common ways to access to the content of slots in S4
objects are either using the function slot(object, name) or
the symbol @ (i.e. object@name). Additional
functions, which are specific for taxlist objects are
taxon_names, taxon_relations,
taxon_traits and taxon_views (see the help
documentation).
Additionally, it is possible to use the methods $ and
[ , the first for access to information in the slot
taxonTraits, while the second can be also used for other
slots in the object.
## acropleustophyte chamaephyte climbing_plant facultative_annual
## 8 25 25 20
## obligate_annual phanerophyte pleustohelophyte reed_plant
## 114 26 8 14
## reptant_plant tussock_plant NA's
## 19 52 35763.3 Subsets
Methods for the function subset are also implemented in
this package. Such subsets usually apply pattern matching (for character
vectors) or logical operations and are analogous to query building in
relational databases. The subset method can be apply to any
slot by setting the value of the argument slot.
Papyrus <- subset(x = Easplist, subset = grepl("papyrus", TaxonName), slot = "names")
summary(Papyrus, "all")Or the very same results:
Papyrus <- subset(x = Easplist, subset = TaxonConceptID == 206, slot = "relations")
summary(Papyrus, "all")Similarly, you can look for a specific name.
3.4 Hierarchical structure
Objects belonging to the class taxlist can optionally
content parent-child relationships and taxonomic levels. Such
information is also included in the data Easplist, as shown
in the summary output.
Easplist## object size: 761.4 Kb
## validation of 'taxlist' object: TRUE
##
## number of taxon usage names: 5393
## number of taxon concepts: 3887
## trait entries: 311
## number of trait variables: 1
## taxon views: 3
##
## concepts with parents: 3698
## concepts with children: 1343
##
## concepts with rank information: 3887
## concepts without rank information: 0
##
## family: 186
## genus: 1011
## complex: 1
## species: 2521
## subspecies: 71
## variety: 95
## form: 2Note that such information can get lost once subset()
has been applied, since the respective parents or children from the
original data set are not anymore in the subset. May you like to recover
parents and children, you can use the functions
get_parents() or get_children(),
respectively.
summary(Papyrus, "all")## ------------------------------
## concept ID: 206
## view ID: 1
## level: species
## parent: none
##
## # accepted name:
## 206 Cyperus papyrus L.
##
## # synonyms (2):
## 52612 Cyperus papyrus ssp. antiquorum (Willd.) Chiov.
## 52613 Cyperus papyrus ssp. nyassicus Chiov.
## ------------------------------
Papyrus <- get_parents(Easplist, Papyrus)
summary(Papyrus, "all")## ------------------------------
## concept ID: 206
## view ID: 1
## level: species
## parent: 54853 Cyperus L.
##
## # accepted name:
## 206 Cyperus papyrus L.
##
## # synonyms (2):
## 52612 Cyperus papyrus ssp. antiquorum (Willd.) Chiov.
## 52613 Cyperus papyrus ssp. nyassicus Chiov.
## ------------------------------
## concept ID: 54853
## view ID: 2
## level: genus
## parent: 55959 Cyperaceae Juss.
##
## # accepted name:
## 54855 Cyperus L.
## ------------------------------
## concept ID: 55959
## view ID: 3
## level: family
## parent: none
##
## # accepted name:
## 55961 Cyperaceae Juss.
## ------------------------------Another way to represent taxonomic ranks is by using the function
indented_list().
indented_list(Papyrus)## Cyperaceae Juss.
## Cyperus L.
## Cyperus papyrus L.4. Applying taxlist to syntaxonomic schemes
4.1 Example of a phytosociological classification
To illustrate the flexibility of the taxlist objects,
the next example will handle a syntaxonomical scheme. As example it will
be used a scheme proposed by the author for aquatic and semi-aquatic
vegetation in Tanzania (Alvarez 2017 Phytocoenologia in
review). The scheme includes 10 associations classified into 4
classes:

4.2 Building the taxlist object
The content for the taxonomic list is included in a data frame and can be downloaded by following command:
load(file.path(path.package("taxlist"), "wetlands_syntax.rda"))The data frame Concepts contains the list of syntaxon
names that are considered as accepted in the previous scheme. This list
will be used to insert the new concepts in the taxlist
object.
head(Concepts)## TaxonConceptID Parent TaxonName
## 1 1 NA Lemnetea minoris
## 2 2 1 Salvinio-Eichhornietalia
## 3 3 2 Pistion stratiotes
## 4 4 3 Lemno paucicostatae-Pistietum stratiotes
## 5 5 NA Potametea
## 6 6 5 Nymphaeetalia loti
## AuthorName Level
## 1 Koch & Tüxen ex den Hartog & Segal 1964 class
## 2 Borhidi ex Borhidi, Muñiz & del Risco 1979 order
## 3 (Schmitz 1971) Schmitz 1988 alliance
## 4 Lebrun 1947 association
## 5 Klika ex Klika & Novák 1941 class
## 6 Lebrun 1947 order
Concepts$TaxonUsageID <- Concepts$TaxonConceptID
Syntax <- df2taxlist(Concepts)## No values for 'AcceptedName' in 'x'. all names will be considered as accepted names.
levels(Syntax) <- c("association", "alliance", "order", "class")
taxon_views(Syntax) <- data.frame(
ViewID = 1, Secundum = "Alvarez (2017)",
Author = "Alvarez M", Year = 2017,
Title = "Classification of aquatic and semi-aquatic vegetation in East Africa",
stringsAsFactors = FALSE
)
Syntax@taxonRelations$ViewID <- 1
Syntax## object size: 11.2 Kb
## validation of 'taxlist' object: TRUE
##
## number of taxon usage names: 26
## number of taxon concepts: 26
## trait entries: 0
## number of trait variables: 0
## taxon views: 1
##
## concepts with parents: 22
## concepts with children: 16
##
## concepts with rank information: 26
## concepts without rank information: 0
##
## class: 4
## order: 5
## alliance: 7
## association: 10Note that the function new created an empty object
(prototype), while levels insert the custom levels
(syntaxonomical hierarchies). For the later function, the levels have to
be inserted from the lower to the higher ranks. Furthermore the
reference defining the concepts included in the syntaxonomic scheme was
inserted in the object using the function taxon_views and
finally the concepts were inserted by the function
add_concept.
The next step will be inserting those names that are considered as
synonyms for the respective syntaxa. Synonyms are included in the data
frame Synonyms.
head(Synonyms)## TaxonConceptID TaxonName
## 1 1 Stratiotetea
## 2 3 Pistion pantropicale
## 3 8 Utriculario-Nymphaeetum
## 4 8 Utriculario exoletae-Nymphaeetum loti
## 5 9 Phragmitetea
## 6 10 Papyretalia
## AuthorName
## 1 den Hartog & Segal 1964
## 2 Schmitz 1971
## 3 (Lebrun 1947) Léonard 1950
## 4 Szafranski & Apema 1983
## 5 Tüxen & Preising 1942
## 6 Lebrun 1947
Syntax <- add_synonym(Syntax,
ConceptID = Synonyms$TaxonConceptID,
TaxonName = Synonyms$TaxonName, AuthorName = Synonyms$AuthorName
)Finally, the codes provided for the associations will be inserted as
traits properties) of them in the slot taxonTraits.
head(Codes)## TaxonConceptID Code
## 1 12 HE1
## 2 13 HE2
## 3 14 HE3
## 4 20 HE4
## 5 17 HE5
## 6 18 HE6
taxon_traits(Syntax) <- Codes
Syntax## object size: 13.8 Kb
## validation of 'taxlist' object: TRUE
##
## number of taxon usage names: 37
## number of taxon concepts: 26
## trait entries: 10
## number of trait variables: 1
## taxon views: 1
##
## concepts with parents: 22
## concepts with children: 16
##
## concepts with rank information: 26
## concepts without rank information: 0
##
## class: 4
## order: 5
## alliance: 7
## association: 10For instance, you may like to get the parental chain from an association (e.g. for Nymphaeetum loti).
Nymplot <- subset(Syntax, charmatch("Nymphaeetum", TaxonName), slot = "names")
summary(Nymplot, "all")## ------------------------------
## concept ID: 8
## view ID: 1
## level: association
## parent: none
##
## # accepted name:
## 8 Nymphaeetum loti Lebrun 1947
##
## # synonyms (2):
## 29 Utriculario-Nymphaeetum (Lebrun 1947) Léonard 1950
## 30 Utriculario exoletae-Nymphaeetum loti Szafranski & Apema 1983
## ------------------------------Note that there is the logical arguments keep_parents
and keep_children to preserve hierarchical information in
the subset:
Nymplot <- subset(Syntax, charmatch("Nymphaeetum", TaxonName),
slot = "names",
keep_parents = TRUE
)
summary(Nymplot, "all")## ------------------------------
## concept ID: 5
## view ID: 1
## level: class
## parent: none
##
## # accepted name:
## 5 Potametea Klika ex Klika & Novák 1941
## ------------------------------
## concept ID: 6
## view ID: 1
## level: order
## parent: 5 Potametea Klika ex Klika & Novák 1941
##
## # accepted name:
## 6 Nymphaeetalia loti Lebrun 1947
## ------------------------------
## concept ID: 7
## view ID: 1
## level: alliance
## parent: 6 Nymphaeetalia loti Lebrun 1947
##
## # accepted name:
## 7 Nymphaeion loti Lebrun 1947
## ------------------------------
## concept ID: 8
## view ID: 1
## level: association
## parent: 7 Nymphaeion loti Lebrun 1947
##
## # accepted name:
## 8 Nymphaeetum loti Lebrun 1947
##
## # synonyms (2):
## 29 Utriculario-Nymphaeetum (Lebrun 1947) Léonard 1950
## 30 Utriculario exoletae-Nymphaeetum loti Szafranski & Apema 1983
## ------------------------------
indented_list(Nymplot)## Potametea Klika ex Klika & Novák 1941
## Nymphaeetalia loti Lebrun 1947
## Nymphaeion loti Lebrun 1947
## Nymphaeetum loti Lebrun 1947By using the function subset() we just created a new
object containing only the association Nymphaeetum loti and its
parental chain. This subset was then used to extract the parental chain
from Syntax.