Efficiency Relative to Storage and Time
Thierry Onkelinx
Source:vignettes/efficiency.Rmd
efficiency.RmdIntroduction
This vignette compares storage and retrieval of data by
git2rdata with other standard R functionality. We consider
write.table() and read.table() for data stored
in a plain text format. saveRDS() and
readRDS() use a compressed binary format.
To get some meaningful results, we will use the nassCDS
dataset from the DAAG
package. We’ll avoid the dependency on the
package by directly downloading the data.
airbag <- read.csv(
"https://vincentarelbundock.github.io/Rdatasets/csv/DAAG/nassCDS.csv"
)
airbag$dead <- airbag$dead == "dead"
airbag$airbag <- airbag$airbag == "airbag"
airbag$seatbelt <- airbag$seatbelt == "belted"
airbag$dvcat <- as.ordered(airbag$dvcat)
str(airbag)
#> 'data.frame': 26217 obs. of 16 variables:
#> $ X : int 1 2 3 4 5 6 7 8 9 10 ...
#> $ dvcat : Ord.factor w/ 5 levels "1-9km/h"<"10-24"<..: 3 2 2 3 3 4 5 5 2 2 ...
#> $ weight : num 25.1 25.1 32.4 495.4 25.1 ...
#> $ dead : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
#> $ airbag : logi FALSE TRUE FALSE TRUE FALSE FALSE ...
#> $ seatbelt : logi TRUE TRUE FALSE TRUE TRUE TRUE ...
#> $ frontal : int 1 1 1 1 1 1 1 1 0 1 ...
#> $ sex : Factor w/ 2 levels "f","m": 1 1 1 1 1 1 2 2 2 1 ...
#> $ ageOFocc : int 26 72 69 53 32 22 22 32 40 18 ...
#> $ yearacc : int 1997 1997 1997 1997 1997 1997 1997 1997 1997 1997 ...
#> $ yearVeh : int 1990 1995 1988 1995 1988 1985 1984 1987 1984 1987 ...
#> $ abcat : Factor w/ 3 levels "deploy","nodeploy",..: 3 1 3 1 3 3 3 3 3 3 ...
#> $ occRole : Factor w/ 2 levels "driver","pass": 1 1 1 1 1 1 1 1 1 1 ...
#> $ deploy : int 0 1 0 1 0 0 0 0 0 0 ...
#> $ injSeverity: int 3 1 4 1 3 3 3 4 1 0 ...
#> $ caseid : Factor w/ 9409 levels "11:1:1","11:1:2",..: 1645 1646 1688 1488 1510 1511 1555 1556 1571 1572 ...Data Storage
On a File System
We start by writing the dataset as is with
write.table(), saveRDS(),
write_vc() and write_vc() without storage
optimization. Note that write_vc() uses optimization by
default. Since write_vc() creates two files for each data
set, we take their combined file size into account.
library(git2rdata)
root <- tempfile("git2rdata-efficient")
dir.create(root)
write.table(airbag, file.path(root, "base_R.tsv"), sep = "\t")
base_size <- file.size(file.path(root, "base_R.tsv"))
saveRDS(airbag, file.path(root, "base_R.rds"))
rds_size <- file.size(file.path(root, "base_R.rds"))
fn <- write_vc(airbag, "airbag_optimize", root, sorting = "X")
#> Warning: `digits` was not set. Setting is automatically to 6. See ?meta
optim_size <- sum(file.size(file.path(root, fn)))
fn <- write_vc(airbag, "airbag_verbose", root, sorting = "X", optimize = FALSE)
#> Warning: `digits` was not set. Setting is automatically to 6. See ?meta
verbose_size <- sum(file.size(file.path(root, fn)))Since the data is highly compressible, saveRDS() yields
the smallest file at the cost of having a binary file format. Both
write_vc() formats yield smaller files than
write.table(). Partly because write_vc()
doesn’t store row names and doesn’t use quotes unless needed. The
difference between the optimized and verbose version of
write_vc() is, in this case, solely due to the way
write_vc() stores factors in the data (tsv)
file. The optimized version stores the indices of the factor whereas the
verbose version stores the levels. For example:
airbag$dvcat has 5 levels with short labels (on average 5
character), storing the index requires 1 character. This results in more
compact files.
| method | file_size | relative |
|---|---|---|
| saveRDS() | 313.13 | 0.12 |
| write_vc(), optimized | 1498.54 | 0.55 |
| write_vc(), verbose | 2225.74 | 0.82 |
| write.table() | 2716.93 | 1.00 |
The reduction in file size when storing in factors depends on the length of the labels, the number of levels and the number of observations. The figure below illustrates the strong gain as soon as the level labels contain more than two characters. The gain is less pronounced when the factor has a large number of levels. The optimization fails in extreme cases with short factor labels and a high number of levels.
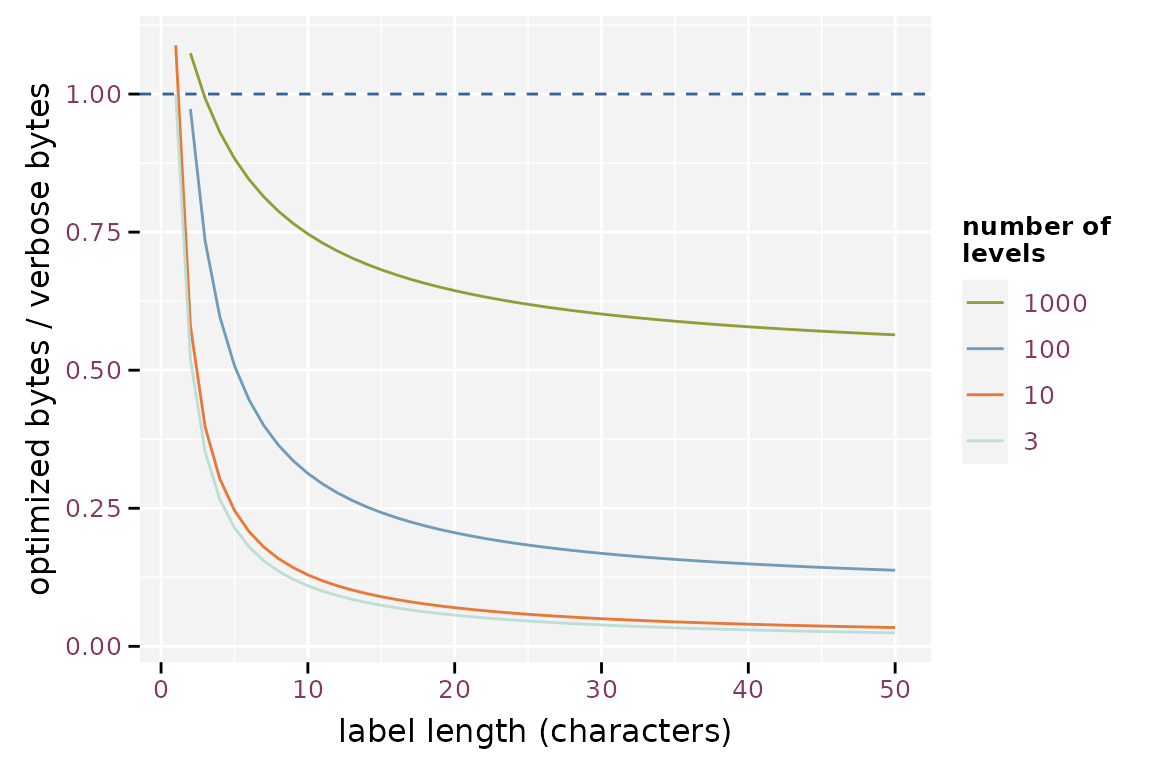
Effect of the label length on the efficiency of storing factor optimized, assuming 1000 observations
The effect of the number of observations is mainly due to the overhead of storing the metadata. The importance of this overhead increases when the number of observations is small.
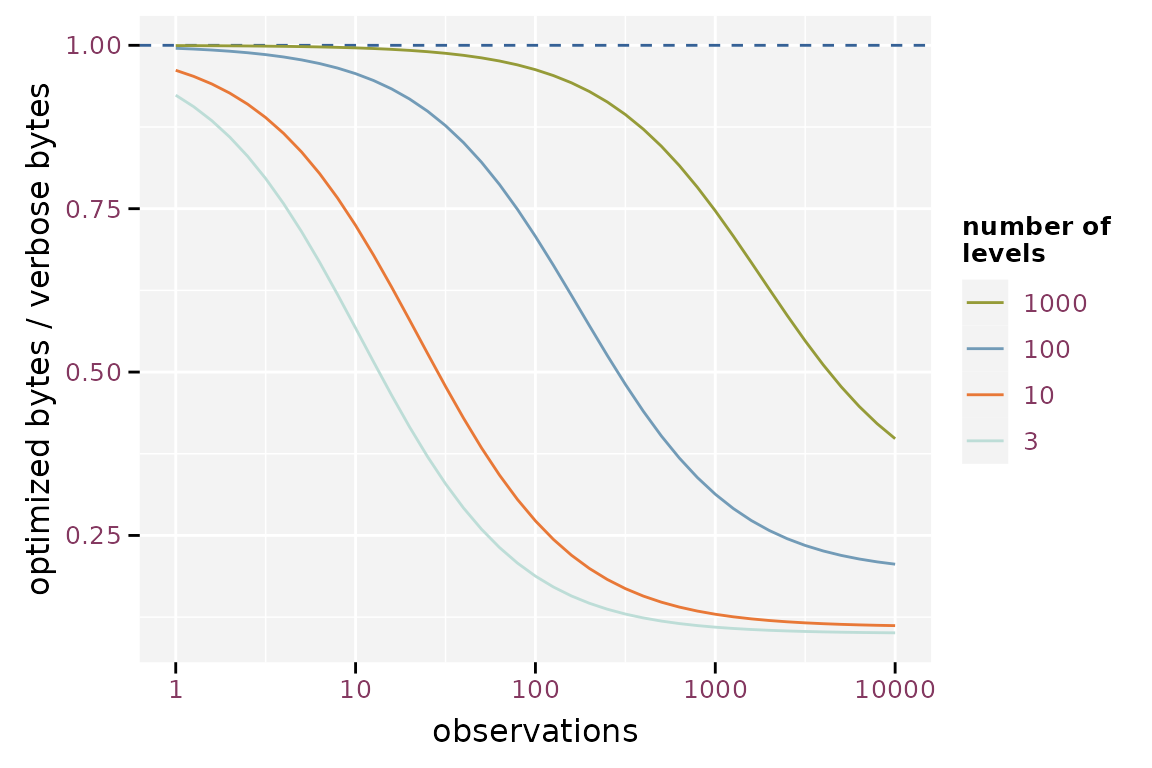
Effect of the number of observations on the efficiency of storing factor optimized assuming labels with 10 characters
In Git Repositories
Here we will simulate how much space the data requires to store the
history in a git repository. We will create a git repository for each
method and store different subsets of the same data. Each commit
contains a new version of the data. Each version is a random sample
containing 90% of the observations of the airbag data. Two
consecutive versions of the subset will have about 90% of the
observations in common.
After writing each version, we commit the file, perform garbage
collection (git gc) on the git repository and then
calculate the size of the git history
(git count-objects -v).
library(git2r)
tmp_repo <- function() {
root <- tempfile("git2rdata-efficient-git")
dir.create(root)
repo <- git2r::init(root)
git2r::config(repo, user.name = "me", user.email = "me@me.com")
return(repo)
}
commit_and_size <- function(repo, filename) {
add(repo, filename)
commit(repo, "test", session = TRUE)
git_size <- system(
sprintf("cd %s\ngit gc\ngit count-objects -v", dirname(repo$path)),
intern = TRUE
)
git_size <- git_size[grep("size-pack", git_size)]
as.integer(gsub(".*: (.*)", "\\1", git_size))
}
repo_wt <- tmp_repo()
repo_wts <- tmp_repo()
repo_rds <- tmp_repo()
repo_wvco <- tmp_repo()
repo_wvcv <- tmp_repo()
repo_size <- replicate(
100,
{
observed_subset <- rbinom(nrow(airbag), size = 1, prob = 0.9) == 1
this <- airbag[
sample(which(observed_subset)),
sample(ncol(airbag))
]
this_sorted <- airbag[observed_subset, ]
fn_wt <- file.path(workdir(repo_wt), "base_R.tsv")
write.table(this, fn_wt, sep = "\t")
fn_wts <- file.path(workdir(repo_wts), "base_R.tsv")
write.table(this_sorted, fn_wts, sep = "\t")
fn_rds <- file.path(workdir(repo_rds), "base_R.rds")
saveRDS(this, fn_rds)
fn_wvco <- write_vc(this, "airbag_optimize", repo_wvco, sorting = "X")
fn_wvcv <- write_vc(
this, "airbag_verbose", repo_wvcv, sorting = "X", optimize = FALSE
)
c(
write.table = commit_and_size(repo_wt, fn_wt),
write.table.sorted = commit_and_size(repo_wts, fn_wts),
saveRDS = commit_and_size(repo_rds, fn_rds),
write_vc.optimized = commit_and_size(repo_wvco, fn_wvco),
write_vc.verbose = commit_and_size(repo_wvcv, fn_wvcv)
)
}
)Each version of the data has on purpose a random order of
observations and variables. This is what would happen in a worst case
scenario as it would generate the largest possible diff. We also test
write.table() with a stable ordering of the observations
and variables.
The randomised write.table() yields the largest git
repository, converging to about 6.5 times the size of a git repository
based on the sorted write.table(). saveRDS()
yields a 26% reduction in repository size compared to the randomised
write.table(), but still is 4.8 times larger than the
sorted write.table(). Note that the gain of storing binary
files in a git repository is much smaller than the gain in individual
file size because git compresses its history. The optimized
write_vc() starts at 88% and converges toward 78%, the
verbose version starts at 94% and converges towards 110%. Storage size
is a lot smaller when using write_vc() with optimization.
The verbose option of write_vc() has little the gain in
storage size. Another advantage is that write_vc() stores
metadata.
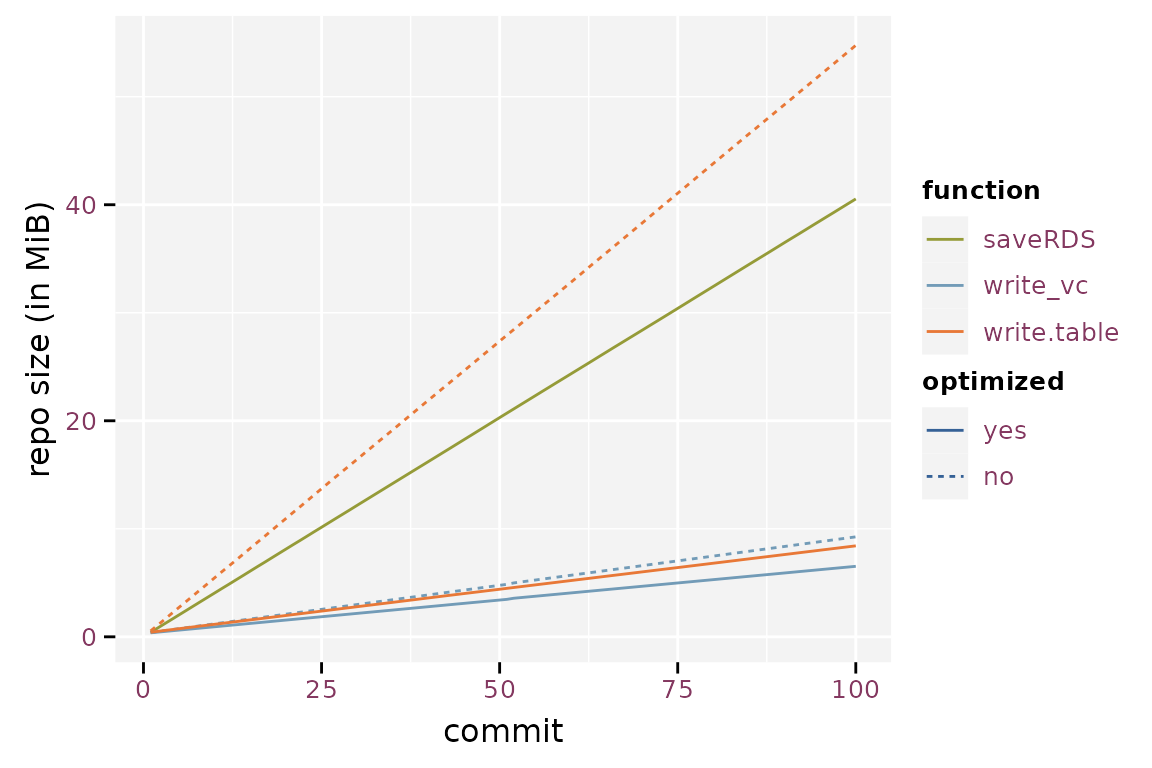
Size of the git history using the different storage methods.
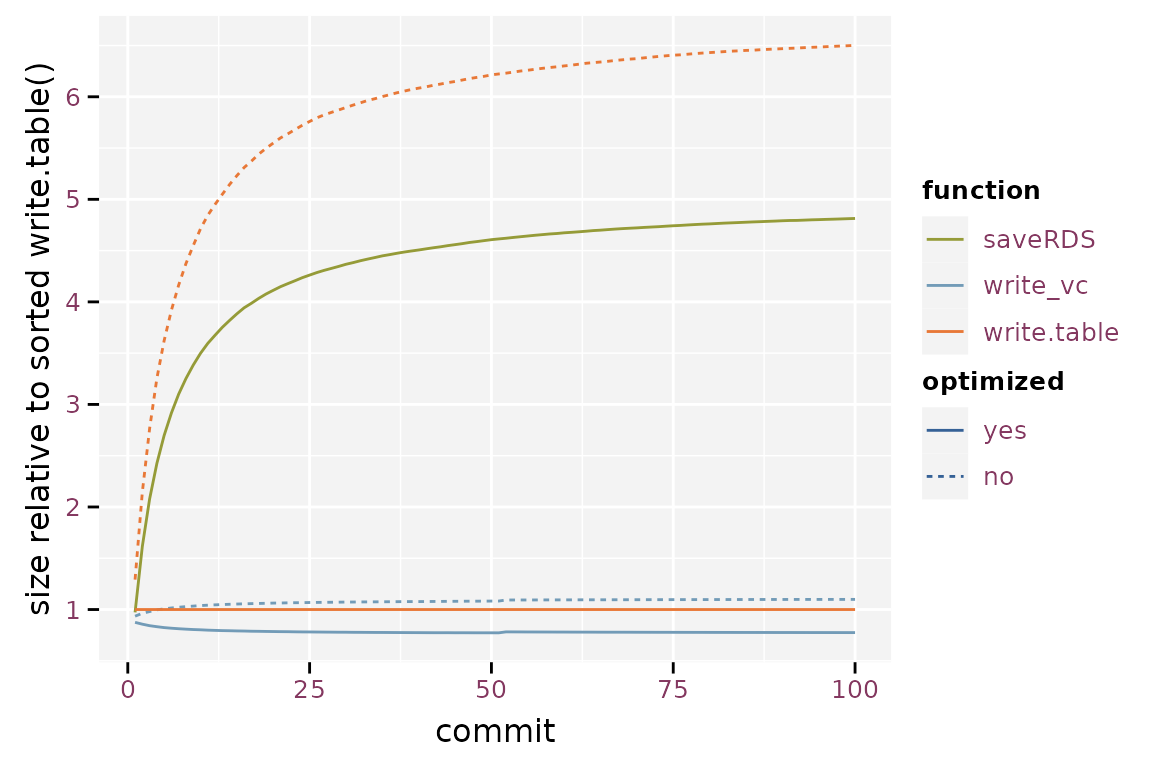
Relative size of the git repository when compared to write.table().
Timings
The code below runs a microbenchmark on the four methods. A microbenchmark runs the code a hundred times and yields a distribution of timings for each expression.
Writing Data
library(microbenchmark)
mb <- microbenchmark(
write.table = write.table(airbag, file.path(root, "base_R.tsv"), sep = "\t"),
saveRDS = saveRDS(airbag, file.path(root, "base_R.rds")),
write_vc.optim = write_vc(airbag, "airbag_optimize", root, sorting = "X"),
write_vc.verbose = write_vc(airbag, "airbag_verbose", root, sorting = "X",
optimize = FALSE)
)
mb$time <- mb$time / 1e6write_vc() takes 155% to 185% more time than
write.table() because it needs to prepare the metadata and
sort the observations and variables. When overwriting existing data,
write_vc() checks the new data against the existing
metadata. saveRDS() requires 53% of the time that
write.table() needs.
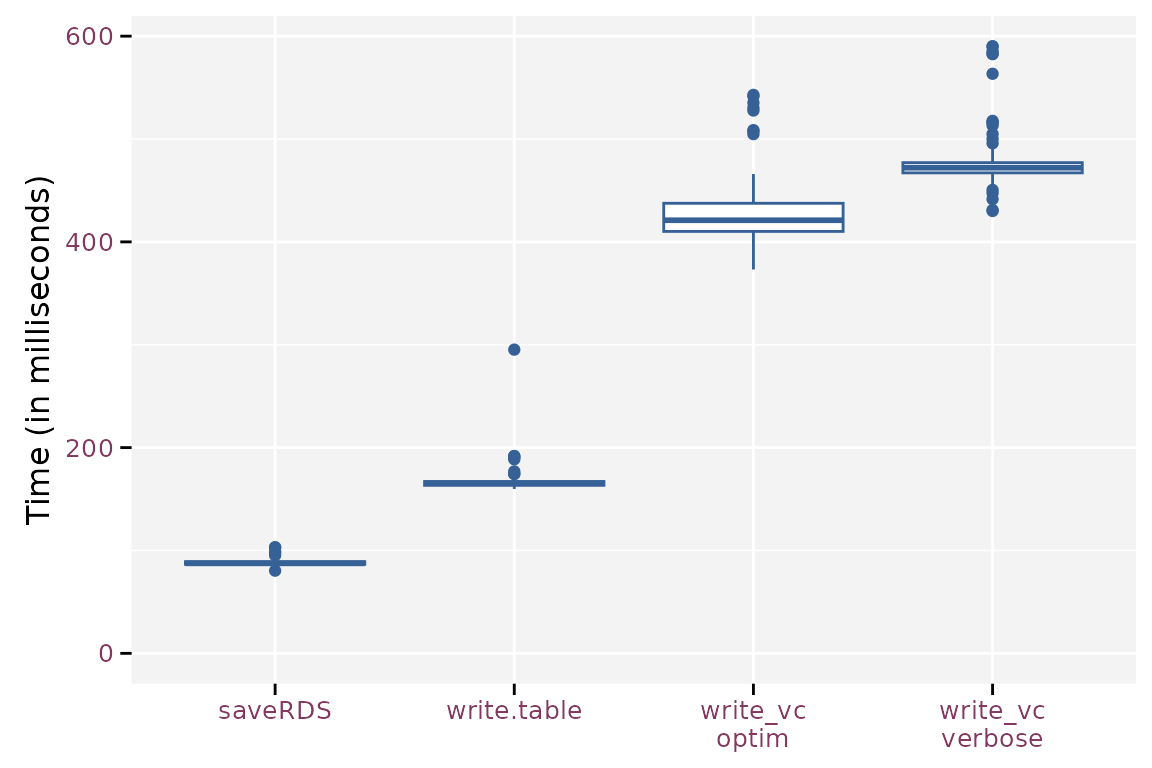
Boxplot of the write timings for the different methods.
Reading Data
mb <- microbenchmark(
read.table = read.table(file.path(root, "base_R.tsv"), header = TRUE,
sep = "\t"),
readRDS = readRDS(file.path(root, "base_R.rds")),
read_vc.optim = read_vc("airbag_optimize", root),
read_vc.verbose = read_vc("airbag_verbose", root)
)
mb$time <- mb$time / 1e6The timings on reading the data is another story. Reading the binary
format takes about 10% of the time needed to read the standard plain
text format using read.table(). read_vc()
takes about 214% (optimized) and 257% (verbose) of the time needed by
read.table(), which at first seems strange because
read_vc() calls read.table() to read the files
and has some extra work to convert the variables to the correct data
type. The main difference is that read_vc() knows the
required data type a priori and passes this info to
read.table(). Otherwise, read.table() has to
guess the correct data type from the file.
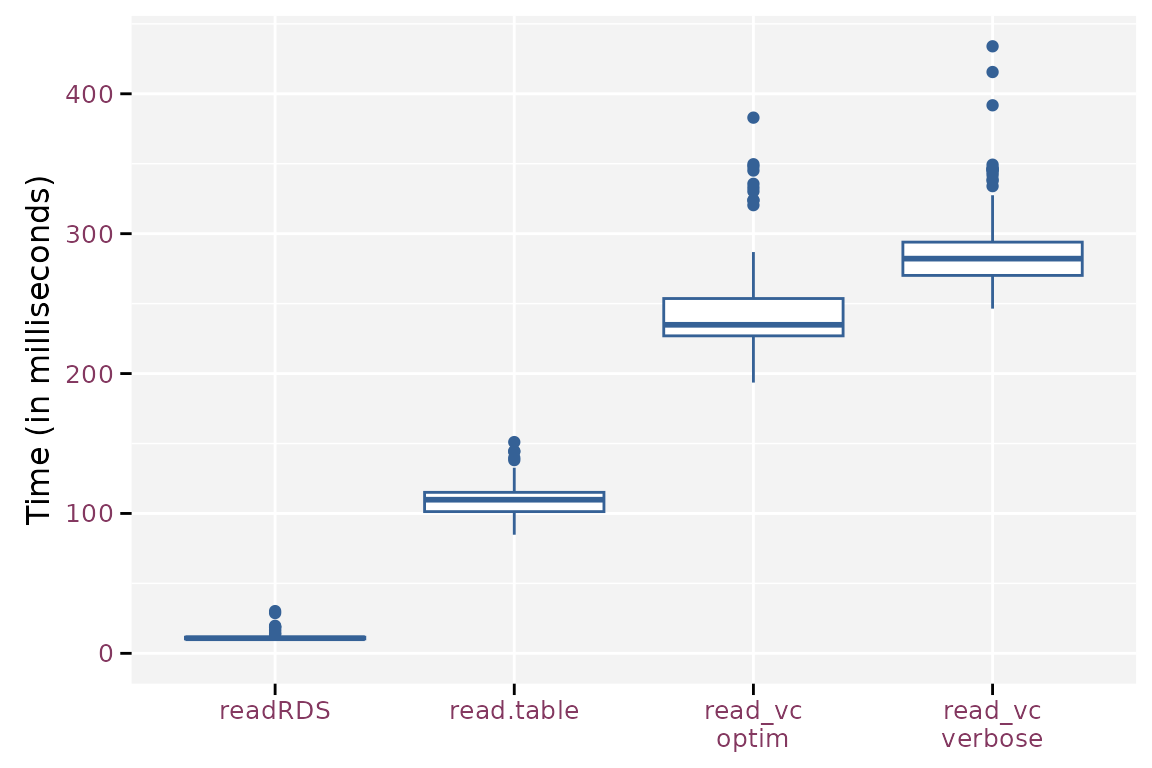
Boxplots for the read timings for the different methods.