
`lingtypology` and other packages
George Moroz
2026-03-03
Source:vignettes/lingtypology_dplyr.Rmd
lingtypology_dplyr.Rmd1. dplyr and pipe |>
It is possible to use dplyr
functions and pipes with lingtypology. It is widely used,
so I will give some examples, how to use it with
thelingtypology package. Using query “list of languages
csv” I found Vincent Garnier’s languages-list
repository. Let’s download and map all the languages from that set.
First download the data:
As we see, some values of the Language.name variable
contain more than one language name. Some of the names probably have
different names in our database. Imagine that we want to map all
languages from Africa. So that the following examples work correctly,
use library(dplyr).
library(dplyr)
new_data |>
mutate(Language.name = gsub(pattern = " ", replacement = "", Language.name)) |>
filter(is.glottolog(Language.name) == TRUE) |>
filter(area.lang(Language.name) == "Africa") |>
select(Language.name) |>
map.feature()We start with a dataframe, here a new_data. First we
remove spaces at the end of each string. Then we check, whether the
language names are in the glottolog database. Then we select only rows
that contain languages of Africa. Then we select the
Language.name variable. And the last line maps all selected
languages.
By default, the values that came from the pipe are treated as the first argument of a function. But when there are some additional arguments, underline sign specify what exact position should be piped to. Let’s produce the same map with a minimap.
new_data |>
mutate(Language.name = gsub(pattern = " ", replacement = "", Language.name)) |>
filter(is.glottolog(Language.name) == TRUE) |>
filter(area.lang(Language.name) == "Africa") |>
select(Language.name) |>
map.feature(languages = _, minimap = TRUE)2. leaflet, leaflet.extras,
mapview, mapedit
There is also a possibility to use lingtypology with
other leaflet
functions (thanks to Niko
Partanen for the idea):
library(leaflet)
map.feature(c("French", "Occitan")) |>
fitBounds(0, 40, 10, 50) |>
addPopups(2, 48, "Great day!")If you add leaflet arguments befor
map.feature function, you need to use argument
pipe.data = _:
leaflet() %>%
fitBounds(0, 40, 10, 50) |>
addPopups(2, 48, "Great day!") |>
map.feature(c("French", "Occitan"), pipe.data = _)The other usage of this pipe.data argument is to put
there a variable with a leaflet object:
m <- leaflet() |>
fitBounds(0, 40, 10, 50) |>
addPopups(2, 48, "Great day!")
map.feature(c("French", "Occitan"), pipe.data = m)If you want to define tiles in leaflet part, you need to
change tile argument in map.feature function, because the
default value for the tile argument is
“OpenStreetMap.Mapnik”.
leaflet() |>
addProviderTiles("Stamen.TonerLite") |>
fitBounds(0, 40, 10, 50) |>
addPopups(2, 48, "Great day!") |>
map.feature(c("French", "Occitan"), pipe.data = _, tile = "none")It is also possible to use some tools provided by leaflet.extras
package:
map.feature(c("French", "Occitan")) |>
leaflet.extras::addDrawToolbar() |>
leaflet.extras::addStyleEditor()
map.feature(c("French", "Occitan")) |>
leaflet.extras::addFullscreenControl()Also there is a nice package mapedit that provide a
possibility of creating and editing of leaflet objects by hand:
map.feature(c("West Circassian", "Russian")) |>
mapedit::editMap() ->
my_polygone
map.feature(c("West Circassian", "Russian")) |>
leaflet::addPolygons(data = my_polygone$finished)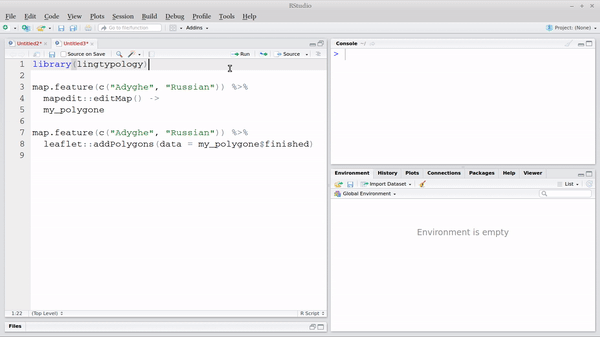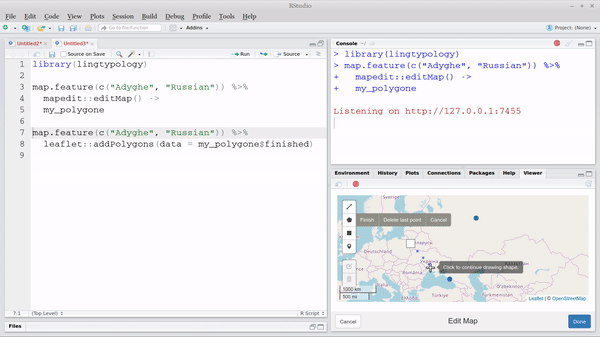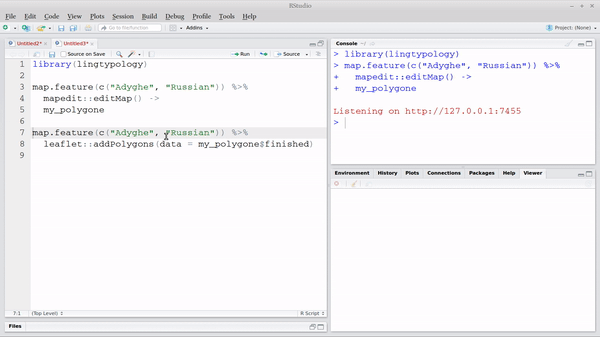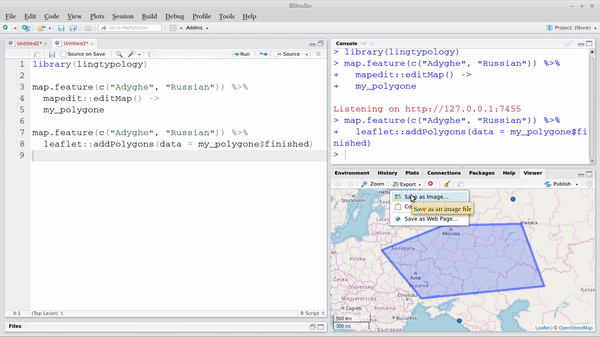
3. Combining maps in a grid and facetisation with
mapview
The leafsync
package provides a possibility to create a multiple maps in a grid
and even synchronise them. There are two functions for that:
latticeview() and sync(). Facetisation is a
really powerfull tool (look for facet_grid() and
facet_wrap() functions from ggplot2).
lingtypology doesn’t provide a facetisation itself, but the
facet argument of the map.feature() function
create a list of maps based on this variable. The result of the work of
this function then is changed: instead of creating a map in Viewer pane
it will return a list that could be used in latticeview()
and sync() functions from the leafsync
package.
faceted <- map.feature(circassian$language,
latitude = circassian$latitude,
longitude = circassian$longitude,
features = circassian$dialect,
facet = circassian$language)
library(leafsync)
sync(faceted, no.initial.sync = FALSE)As you can see we provided a circassian$language to the
facet argument, so it returned a list of two maps that
stored in faceted variable.
It is also possible to combine any maps that were created, just store
them in a variable, and combine them in latticeview() and
sync() functions
m1 <- map.feature(lang.aff("Tsezic"), label = lang.aff("Tsezic"))
m2 <- map.feature(lang.aff("Avar-Andi"), label = lang.aff("Avar-Andi"))
sync(m1, m2)4. Get data from OpenStreetMap with overpass
This section is inspired by talk with Niko Partanen and his gist. Overpass is a packge with tools to work with the OpenStreetMap (OSM) Overpass API. Explore simple Overpass queries with overpass turbo. Imagine that we need to get all settlements from Ingushetia, Daghestan and Chechnya. So, first, load a library:
library(overpass)Create a query:
settlements <- 'area[name~"Дагестан|Ингушетия|Чечня"];
(node["place"~"city|village|town|hamlet"](area););
out;'Pass the query to overpass_query() function and change
the input result to dataframe:
query_result <- overpass_query(settlements)
settlement_data <- as.data.frame(query_result[, c("id", "lon", "lat", "name")])Some values could be NA, so I profer clean it with
complete.cases() function:
settlement_data <- settlement_data[complete.cases(settlement_data),]On the last step, I will use a “fake” language argument to avoid the creation of some Glottolog links:
map.feature(language = "fake",
latitude = settlement_data$lat,
longitude = settlement_data$lon,
label = settlement_data$name)Results are not ideal: there are some villages Дагестанская and
Красный Дагестан in Adygeya and Krasnodarskiy district, but the most
points are correct. It is also possible to get all data from some
polygone created with mapedit (see previous section).
5. Create your own atlas with rmarkdown
This section is inspired by talk with Niko Partanen. It is possible
to create an atlas website using lingtypology and rmarkdown
packages. The function atlas.database() creates a folder in
the working directory that contains an rmarkdown template
for a web-site.
First, lets create a dataframe with some data.
df <- wals.feature(c("1a", "20a"))Second we can create a website using atlas.database()
function:
-
languagesargument is a language list -
featuresargument is a data.frame with corresponding features -
latitudeandlongitudearguments are optional
atlas.database(languages = df$language,
features = df[,c(4:5)],
latitude = df$latitude,
longitude = df$longitude,
atlas.name = "Some WALS features",
author = "Author Name")We can see that this function creates a subfolder with following files:
list.files("./atlas_Some_WALS_features/")The last step is to run a command:
rmarkdown::render_site("./atlas_Some_WALS_features/")Then the atlas website will be created (here is a result). If you want to change something in the website, just change some files:
- write information about atlas in index.Rmd file
- list citation information
- change any
.Rmdfile - …
- and on the end rerun the
rmarkdown::render_site("./atlas_Some_WALS_features/")command.
6. Create .kml file using sp and
rgdal
.kml file is a common file type for geospatial data. This kind of
files are used in Google Earth, Gabmap (a web application
that visualizes dialect variations) and others. In order to produce a
.kml file you need to have a dataset with coordinates such as
circassian:
sp::coordinates(circassian) <- ~longitude+latitude
sp::proj4string(circassian) <- sp::CRS("+proj=longlat +datum=WGS84")
rgdal::writeOGR(circassian["village"],
"circassian.kml",
layer="village",
driver="KML")