
Get started with rmangal
Steve Vissault
Kevin Cazelles
2025-11-11
Source:vignettes/rmangal.Rmd
rmangal.RmdContext
The Mangal project
The Mangal project aims at archiving published ecological networks and at easing their retrieval. To do so, Mangal:
uses a data specification for ecological networks (described in Poisot et al. 2016);
archives ecological networks in a PostgreSQL database;
provides:
- a data explorer to visualize and download data available;
- a RESTful Application Programming Interface (API);
- a client library for Julia: Mangal.jl;
- a client of this API for R: the rmangal package described below.
Currently, 175 datasets are included in the database representing over 1300 ecological networks. In 2016, the first paper describing the project was published and introduced the first release of rmangal (Poisot et al. 2016). Since then, the structure of the database has been improved (new fields have been added), several ecological networks have been added and the API entirely rewritten. Consequently, the first release of the rmangal is obsolete (and archived) and we introduce rmangal v2.0 in this vignette. This vignette was reviewed on November 2025, when version 2.2 was released.
Data structure
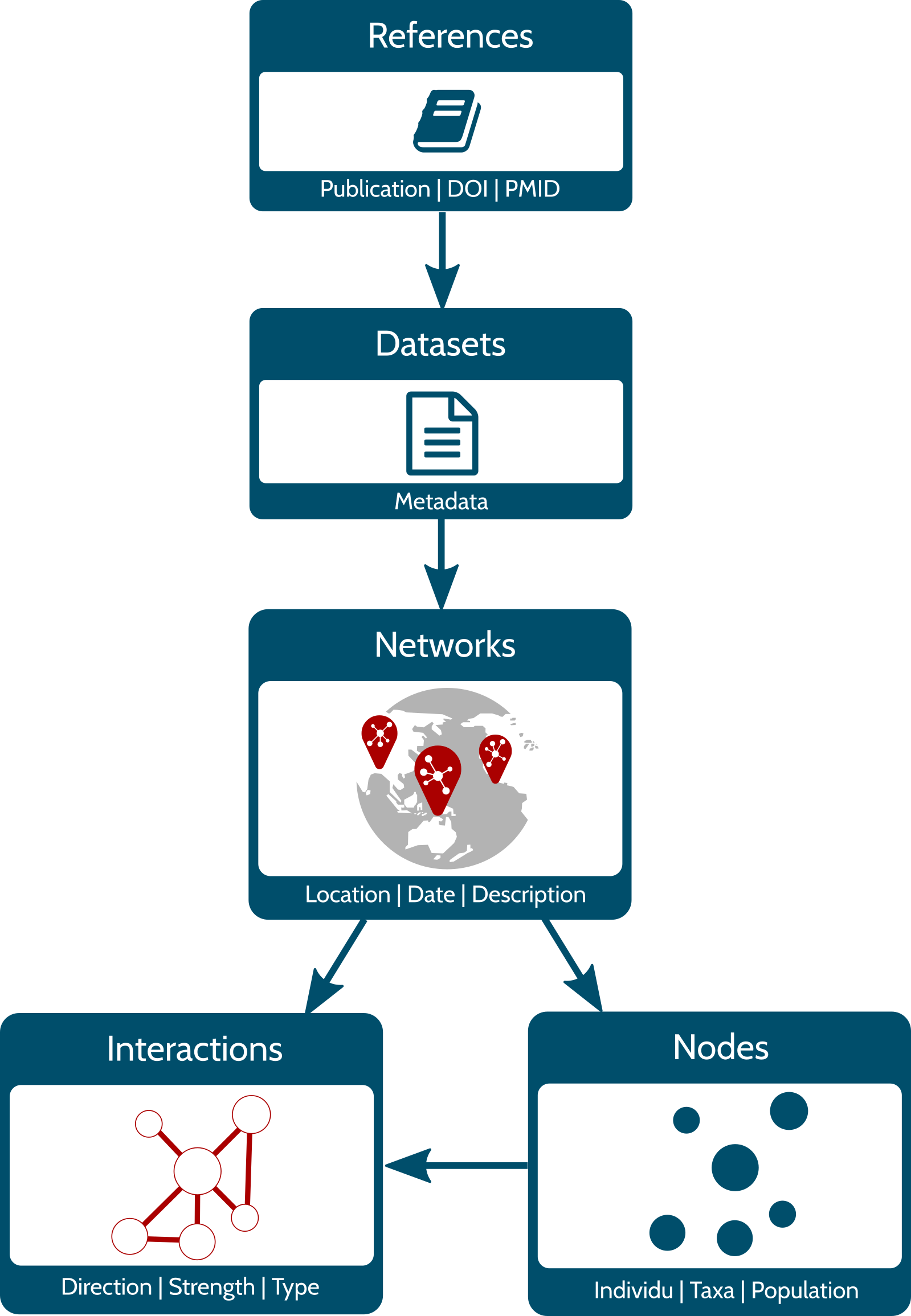
The diagram on the left side represents the structure of the Mangal database. All references included in Mangal correspond to a specific publication that includes one or several dataset(s). This dataset is basically a collection of ecological networks whose nodes and interactions (edges) are stored in separate tables. Below, we briefly describe the content of each table.
References – Information pertaining to a reference
(scientific article, book, online website, etc.) characterizing an
original collection of ecological networks. URLs of data and publication
sources are included as well as persistent identifiers (when available)
such as digital object identifiers (DOIs). This allows users to retrieve
more details about original publications using appropriate R packages
such as crossref.
Datasets – Metadata of the datasets attached to a reference. It includes a general description of the networks.
Networks – Metadata of the networks attached to a dataset. It provides the sampling location, date and specific description of the network.
Nodes – Information on the population, taxa or
individual in the network. Each node has the original taxon name
documented and a taxonomic backbone provided by all services embedded in
taxize (Chamberlain et al.
2019).
Interactions – Information on the interaction type (e.g. mutualism, predation, etc.), the strength, and the direction of the interaction between two nodes.
Authentication
So far, the rmangal package provides methods to get
access to the data store. Data requests (performed via
rmangal_request() or
rmangal_request_singleton()) do not require any
authentication.
A bearer authentication strategy using ORCID credentials (as a third-party
services) has been implemented on all POST,
DELETE, PUT API operations to allow the user
to add and delete new ecological to the database. These features are not
currently included in the rmangal package, but remain
under consideration for future major releases.
How to use rmangal
Overall approach
In order to efficiently retrieve networks from the database, rmangal includes 7 search functions querying the 5 tables described above as well as a table dedicated to the taxonomy backbone.
-
search_references(): search in the reference table, for instance the user can look for a specificdoi; -
search_datasets(): search among datasets using a keyword; -
search_networks()andsearch_networks_sf(): search networks based on a keyword or a geographical area; -
search_interactions(): list all networks containing a specific interaction type; -
search_nodes(): identify nodes based on nodes information; -
search_taxonomy(): identify nodes based on taxonomic names and unique identifiers.
All of these functions return specific class objects with the
information needed to retrieve the corresponding set of ecological
networks with get_collection(). Hence, the user can easily
retrieve data in two steps:
networks <- search_*(query = "your query") |> get_collection()If there is only one network to be retrieved,
get_collection() returns a mgNetwork object,
otherwise it returns an object of class
mgNetworksCollection which is a collection (a list) of
mgNetwork objects. Below, we exemplify how to use the
search functions, how to get a collection of networks and how to use
other packages to carry out specific analyses.
Requests can be cached (rmangal leverages
httr2::req_cache()) either by setting the argument
cache to TRUE or by providing a path that
points to the desired cache directory. Using cache the
basic workflow becomes:
Search functions
In rmangal, every function queries a specific table and allows only one query at a time (see section Batch analysis to learn how to perform more than one query). All the functions offer two ways to query the corresponding table:
- a keyword: in this case, the entries returned are the partial or full keyword match of any strings contained in the table;
- a custom query: in this case, entries returned are exact matches.
We start by loading rmangal as well as
tibble (enhanced data frames).
Search and list available datasets
Let’s assume we are looking for ecological networks including species
living in lagoons. If we have no idea about any existing data set, the
best starting point is then to query the dataset table with
lagoon as a keyword:
lagoon <- search_datasets(query = "lagoon")
class(lagoon)
#> [1] "tbl_df" "tbl" "data.frame" "mgSearchDatasets"
lagoon
#> # A tibble: 2 × 10
#> id name description public created_at updated_at ref_id user_id networks references
#> <int> <chr> <chr> <lgl> <chr> <chr> <int> <int> <list> <list>
#> 1 22 zetina_2003 Dietary matrix of the Huizache–Caimanero lagoon TRUE 2019-02-23T17:04:32.0… 2019-02-2… 22 3 <df> <df>
#> 2 52 yanez_1978 Food web of the Guerrero lagoon TRUE 2019-02-24T23:42:52.0… 2019-02-2… 53 3 <df> <df>If the Mangal reference identifiers of this dataset is known, the following query can be used:
lagoon_zetina <- search_datasets(list(ref_id = 22))
lagoon_zetina
#> # A tibble: 1 × 10
#> id name description public created_at updated_at ref_id user_id networks references
#> <int> <chr> <chr> <lgl> <chr> <chr> <int> <int> <list> <list>
#> 1 22 zetina_2003 Dietary matrix of the Huizache–Caimanero lagoon TRUE 2019-02-23T17:04:32.0… 2019-02-2… 22 3 <df> <df>Note that if an empty character is passed, i.e. "", all
entries are returned.
all_datasets <- search_datasets("")
head(all_datasets)
#> # A tibble: 6 × 10
#> id name description public created_at updated_at ref_id user_id networks references
#> <int> <chr> <chr> <lgl> <chr> <chr> <int> <int> <list> <list>
#> 1 2 howking_1968 Insect activity recorded on flower at Lake Hazen… TRUE 2019-02-2… 2019-02-2… 2 2 <df> <df>
#> 2 7 lundgren_olesen_2005 Pollnator activity recorded on flowers, Uummanna… TRUE 2019-02-2… 2019-02-2… 7 2 <df> <df>
#> 3 9 elberling_olesen_1999 Flower-visiting insect at Mt. Latnjatjarro, nort… TRUE 2019-02-2… 2019-02-2… 9 2 <df> <df>
#> 4 14 johnston_1956 Predation by short-eared owls on a salicornia sa… TRUE 2019-02-2… 2019-02-2… 14 3 <df> <df>
#> 5 15 havens_1992 Pelagic communities of small lakes and ponds of … TRUE 2019-02-2… 2019-02-2… 15 3 <df> <df>
#> 6 16 kemp_1977 Food web for the Crystal River estuary TRUE 2019-02-2… 2019-02-2… 16 3 <df> <df>
length(all_datasets)
#> [1] 10As shown in the diagram above, a dataset comes from a specific
reference and search_references() queries the reference
table directly. A handy argument of this function is doi as
it allows to pass a Digital Object Identifier and so to retrieve all
datasets attached to a specific publication.
zetina_2003 <- search_references(doi = "10.1016/s0272-7714(02)00410-9")Finding a specific network
We can also search by keywords across all networks.
insect_coll <- search_networks(query = "insect%")
head(insect_coll)
#> id name date
#> 1 18 mosquin_martin_1967_19650731_18 1965-07-31T00:00:00.000Z
#> 2 909 elberling_olesen_1999_19940823_909 1994-08-23T00:00:00.000Z
#> 3 948 kato_1993_19910901_948 1991-09-01T00:00:00.000Z
#> 4 1460 cornaby_1974_19680208_1460 1968-02-08T00:00:00.000Z
#> 5 1461 cornaby_1974_19680229_1461 1968-02-29T00:00:00.000Z
#> 6 1471 jiron_cartin_1981_19770101_1471 1977-01-01T00:00:00.000Z
#> description
#> 1 Occurence of flower-visiting insect on plant species, two miles north of Bailey Point, Melville Island, N.W.T., Canada
#> 2 Flower-visiting insect at Mt. Latnjatjarro, northern Sweden
#> 3 Flower and anthophilous insect interactions in the primary cool-temperate subalpine forests and meadows at Mt. Kushigata, Yamanashi Prefecture, Japan
#> 4 The insect community of a toad carrion in a tropical dry lowland forest at Finac La pacifica, Guanacaste Prov., Costa Rica
#> 5 The insect community of a toad carrion in a tropical wet lowland forest near Rincon de Osa, Puntarenas Prov., Costa Rica
#> 6 The insect community of a dog carcass in a premontane humid forest, University of Costa Rica, Costa Rica
#> public all_interactions created_at updated_at dataset_id user_id geom_type geom_lon geom_lat
#> 1 TRUE FALSE 2019-02-22T18:38:37.491Z 2019-02-22T18:38:37.491Z 4 3 Point -114.9667 75
#> 2 TRUE FALSE 2019-02-24T22:21:32.444Z 2019-02-24T22:21:32.444Z 9 2 Point 18.5 68.35
#> 3 TRUE FALSE 2019-02-25T20:52:09.499Z 2019-02-25T20:52:09.499Z 66 2 Point 138.3833 35.5833
#> 4 TRUE FALSE 2019-03-01T18:30:50.890Z 2019-03-01T18:30:50.890Z 91 4 Point -85.09443 10.4568
#> 5 TRUE FALSE 2019-03-01T18:30:57.419Z 2019-03-01T18:30:57.419Z 91 4 Point -83.50833 8.534018
#> 6 TRUE FALSE 2019-03-04T18:22:33.907Z 2019-03-04T18:22:33.907Z 99 4 Point -84.07651 9.933982It is also possible to retrieve all networks based on interaction types involved:
comp_interac <- search_interactions(type = "competition")
# Number of competition interactions in mangal
nrow(comp_interac)
#> [1] 12search_networks_sf() handles spatial queries: argument
query_sf takes a sf object
as input and returns all networks included in the spatial extent of this
object. For instance, one can retrieve all networks found in North
Carolina included in Mangal like so:
library(sf)
library(mapview)
area <- sf::st_read(system.file("shape/nc.shp", package="sf"))
#> Reading layer `nc' from data source `/usr/lib/R/site-library/sf/shape/nc.shp' using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> Geodetic CRS: NAD27
in_NC <- search_networks_sf(area)
mapView(
st_geometry(area),
color = "red",
legend = FALSE,
col.regions = "#FF000033"
) + mapView(in_NC, legend = FALSE) 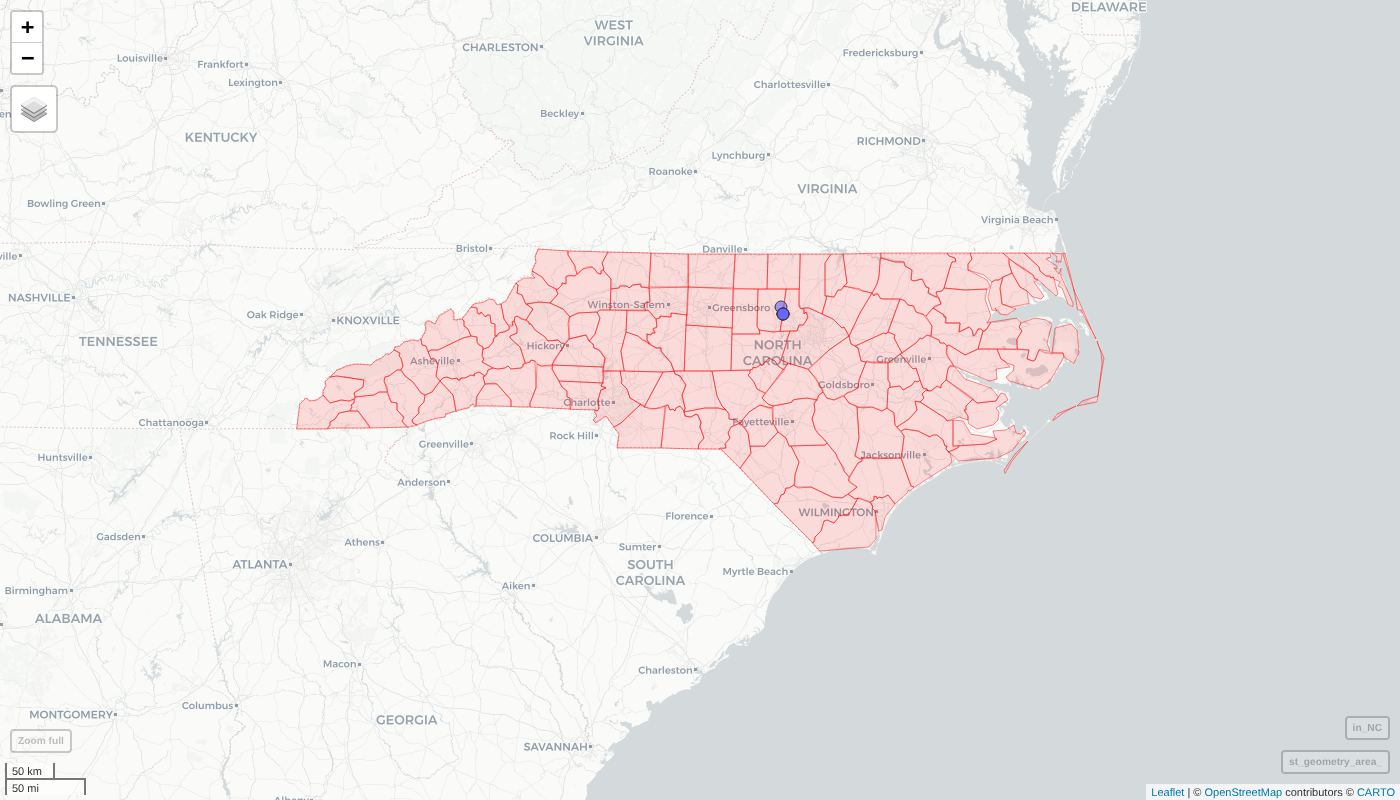
Map of the networks found in North Carolina
Search for a specific taxon
The user can easily identify networks including a specific taxonomic
entity with search_taxonomy():
sr_ficus <- search_taxonomy("Ficus")This function allows to search for a specific taxonomic entity with
its validated name or unique identifiers, i.e. EOL, TSN, GBIF, COL, BOLD
and NCBI IDs. Taxon names of the taxonomy table were
validated with TNRS (see https://tnrs.biendata.org/) and/or GNR (see https://resolver.globalnames.org/). The taxon names in
this table might not be the taxon name documented in the original
publication. In order to identify relevant networks with the original
name, use search_nodes().
The validation of taxon names was performed by an automated procedure
using taxize (Chamberlain et al.
2019) and if there is any doubt, the original names recorded by
authors should be regarded as the most reliable information. Please
report any issue related to taxonomy at https://github.com/mangal-interactions/contribute/issues/new/choose.
head(search_taxonomy(tsn = 28749))
#> id original_name node_level network_id taxonomy_id created_at updated_at taxonomy.id taxonomy.name
#> 1 2629 Acer negundo taxon 19 2 2019-02-22T18:48:49.433Z 2019-02-22T18:48:49.433Z 2 Acer negundo
#> taxonomy.ncbi taxonomy.tsn taxonomy.eol taxonomy.bold taxonomy.gbif taxonomy.col taxonomy.rank taxonomy.created_at
#> 1 4023 28749 583069 100987 3189866 90203e29e2f59e5754167f89b9eba3cc species 2019-02-21T21:17:12.585Z
#> taxonomy.updated_at
#> 1 2019-06-14T15:20:36.273Z
head(search_taxonomy(eol = 583069))
#> id original_name node_level network_id taxonomy_id created_at updated_at taxonomy.id taxonomy.name
#> 1 2629 Acer negundo taxon 19 2 2019-02-22T18:48:49.433Z 2019-02-22T18:48:49.433Z 2 Acer negundo
#> taxonomy.ncbi taxonomy.tsn taxonomy.eol taxonomy.bold taxonomy.gbif taxonomy.col taxonomy.rank taxonomy.created_at
#> 1 4023 28749 583069 100987 3189866 90203e29e2f59e5754167f89b9eba3cc species 2019-02-21T21:17:12.585Z
#> taxonomy.updated_at
#> 1 2019-06-14T15:20:36.273ZNote that in some cases, it may be necessary to locate a dataset
using the original name provided in the publication; in such cases, the
search_nodes() function should be used.
sr_ficus2 <- search_nodes("Ficus")Get networks associated with a search_* object
Once the search performed, ecological networks are accessible from
the object returned with get_collection():
nets_lagoons <- lagoon |> get_collection()
nets_in_NC <- in_NC |> get_collection()
nets_competition <- comp_interac |> get_collection()
nets_lagoons
class(nets_lagoons)
#> [1] "mgNetworksCollection"Note that mgNetworksCollection objects are lists of
mgNetwork object which are a list of five datasets
reflecting the 5 tables presented in the diagram in the first
section:
names(nets_lagoons[[1]])
#> [1] "network" "nodes" "interactions" "dataset" "reference"
head(nets_lagoons[[1]]$network)
#> network_id name date description public all_interactions
#> 1 86 zetina_2003_20030101_86 2003-01-01T00:00:00.000Z Dietary matrix of the Huizache–Caimanero lagoon TRUE FALSE
#> created_at updated_at dataset_id user_id geom_type geom_lon geom_lat
#> 1 2019-02-23T17:04:34.046Z 2019-02-23T17:04:34.046Z 22 3 Point -106.1099 22.98531
head(nets_lagoons[[1]]$nodes)
#> node_id original_name node_level network_id taxonomy_id created_at updated_at taxonomy.id taxonomy.name
#> 1 4904 Scianids taxon 86 4363 2019-02-23T17:04:42.505Z 2019-02-23T17:04:42.505Z 4363 Sciaenidae
#> 2 4905 Elopids taxon 86 4364 2019-02-23T17:04:42.571Z 2019-02-23T17:04:42.571Z 4364 Elops
#> 3 4906 Lutjanids taxon 86 4365 2019-02-23T17:04:42.622Z 2019-02-23T17:04:42.622Z 4365 Lutjanidae
#> 4 4907 Carangids taxon 86 4366 2019-02-23T17:04:42.672Z 2019-02-23T17:04:42.672Z 4366 Carangidae
#> 5 4908 Centropomids taxon 86 4367 2019-02-23T17:04:42.728Z 2019-02-23T17:04:42.728Z 4367 Centropomidae
#> 6 4909 Ariids taxon 86 4368 2019-02-23T17:04:42.786Z 2019-02-23T17:04:42.786Z 4368 Ariidae
#> taxonomy.ncbi taxonomy.tsn taxonomy.eol taxonomy.bold taxonomy.gbif taxonomy.col taxonomy.rank taxonomy.created_at
#> 1 30870 169237 5211 1856 NA 81a86c329909d507edb5c296906ef3f4 family 2019-02-23T17:04:35.620Z
#> 2 7927 28630 46561210 4061 NA 94532a14786adeb25bcec244a53aadc1 genus 2019-02-23T17:04:35.744Z
#> 3 30850 168845 5294 1858 NA 7150078b7dd31a5f7575240f1b76f834 family 2019-02-23T17:04:35.870Z
#> 4 8157 168584 5361 1851 NA 1ccc9e80931658b72d166c1764b687b5 family 2019-02-23T17:04:35.975Z
#> 5 8184 167642 5355 586 NA 529f1f934910702cb5334f8aa90cd22f family 2019-02-23T17:04:36.102Z
#> 6 31017 43998 5115 1313 NA f60963ef9a967267b989ec22096edd3b family 2019-02-23T17:04:36.207Z
#> taxonomy.updated_at taxonomy
#> 1 2019-06-14T15:25:46.438Z NA
#> 2 2019-06-14T15:25:46.492Z NA
#> 3 2019-06-14T15:25:46.546Z NA
#> 4 2019-06-14T15:25:46.600Z NA
#> 5 2019-06-14T15:25:46.654Z NA
#> 6 2019-06-14T15:25:46.708Z NA
head(nets_lagoons[[1]]$interactions)
#> interaction_id node_from node_to date direction type method attr_id value geom public network_id
#> 1 48376 4912 4912 2003-01-01T00:00:00.000Z directed predation NA 12 0.026 NA TRUE 86
#> 2 48377 4912 4914 2003-01-01T00:00:00.000Z directed predation NA 12 0.025 NA TRUE 86
#> 3 48378 4912 4915 2003-01-01T00:00:00.000Z directed predation NA 12 0.003 NA TRUE 86
#> 4 48379 4912 4918 2003-01-01T00:00:00.000Z directed predation NA 12 0.009 NA TRUE 86
#> 5 48380 4912 4919 2003-01-01T00:00:00.000Z directed predation NA 12 0.009 NA TRUE 86
#> 6 48381 4912 4920 2003-01-01T00:00:00.000Z directed predation NA 12 0.016 NA TRUE 86
#> created_at updated_at attribute.id attribute.name attribute.description
#> 1 2019-02-23T17:05:45.061Z 2019-02-23T17:05:45.061Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> 2 2019-02-23T17:05:45.131Z 2019-02-23T17:05:45.131Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> 3 2019-02-23T17:05:45.193Z 2019-02-23T17:05:45.193Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> 4 2019-02-23T17:05:45.247Z 2019-02-23T17:05:45.247Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> 5 2019-02-23T17:05:45.309Z 2019-02-23T17:05:45.309Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> 6 2019-02-23T17:05:45.367Z 2019-02-23T17:05:45.367Z 12 dietary matrix Proportions of the consumer diets made up by the prey.
#> attribute.unit attribute.created_at attribute.updated_at
#> 1 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
#> 2 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
#> 3 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
#> 4 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
#> 5 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
#> 6 NA 2019-02-23T17:04:25.350Z 2019-02-23T17:04:25.350Z
head(nets_lagoons[[1]]$dataset)
#> dataset_id name description public created_at updated_at ref_id
#> 1 22 zetina_2003 Dietary matrix of the Huizache–Caimanero lagoon TRUE 2019-02-23T17:04:32.017Z 2019-02-23T17:04:32.017Z 22
#> user_id
#> 1 3
head(nets_lagoons[[1]]$reference)
#> ref_id doi first_author year jstor pmid
#> 1 22 10.1016/s0272-7714(02)00410-9 manuel j. zetina-rejon 2003 NA NA
#> bibtex
#> 1 @article{Zetina_Rej_n_2003, doi = {10.1016/s0272-7714(02)00410-9}, url = {https://doi.org/10.1016%2Fs0272-7714%2802%2900410-9}, year = 2003, month = {aug}, publisher = {Elsevier {BV}}, volume = {57}, number = {5-6}, pages = {803--815}, author = {Manuel J. Zetina-Rejón and Francisco Arreguí-Sánchez and Ernesto A. Chávez}, title = {Trophic structure and flows of energy in the Huizache{\textendash}Caimanero lagoon complex on the Pacific coast of Mexico},journal = {Estuarine, Coastal and Shelf Science}}
#> paper_url data_url created_at updated_at
#> 1 https://doi.org/10.1016%2Fs0272-7714%2802%2900410-9 https://globalwebdb.com/ 2019-02-23T17:04:28.307Z 2019-02-23T17:04:28.307ZIntegrated workflow with rmangal
Batch analysis
So far, the search functions of rmangal allow the
user to perform only a single search at a time. The simplest way to do
more than one search is to loop over a vector or a list of queries.
Below we exemplify how to do so using lapply():
tsn <- c(837855, 169237)
mgn <- lapply(tsn, function(x) search_taxonomy(tsn = x)) |>
lapply(get_collection) |>
combine_mgNetworks()
mgnGeolocate Mangal networks with sf
The function get_collection() has an argument
as_sf than converts network metadata of mgNetwork objects
to sf objects, which requires sf to be
installed. This allows the user to easily track the location of the
networks retrieved from Mangal.
# assuming sf and mapview are is loaded (as we did above)
mg_lag_sf <- search_datasets(query = 'lagoon') |>
get_collection(as_sf = TRUE)
class(mg_lag_sf[[1]]$network)
#> [1] "sf" "data.frame"
# let's combine all these sf object into a single one
mapView(mg_lag_sf[[1]]$network) + mapView(mg_lag_sf[[2]]$network)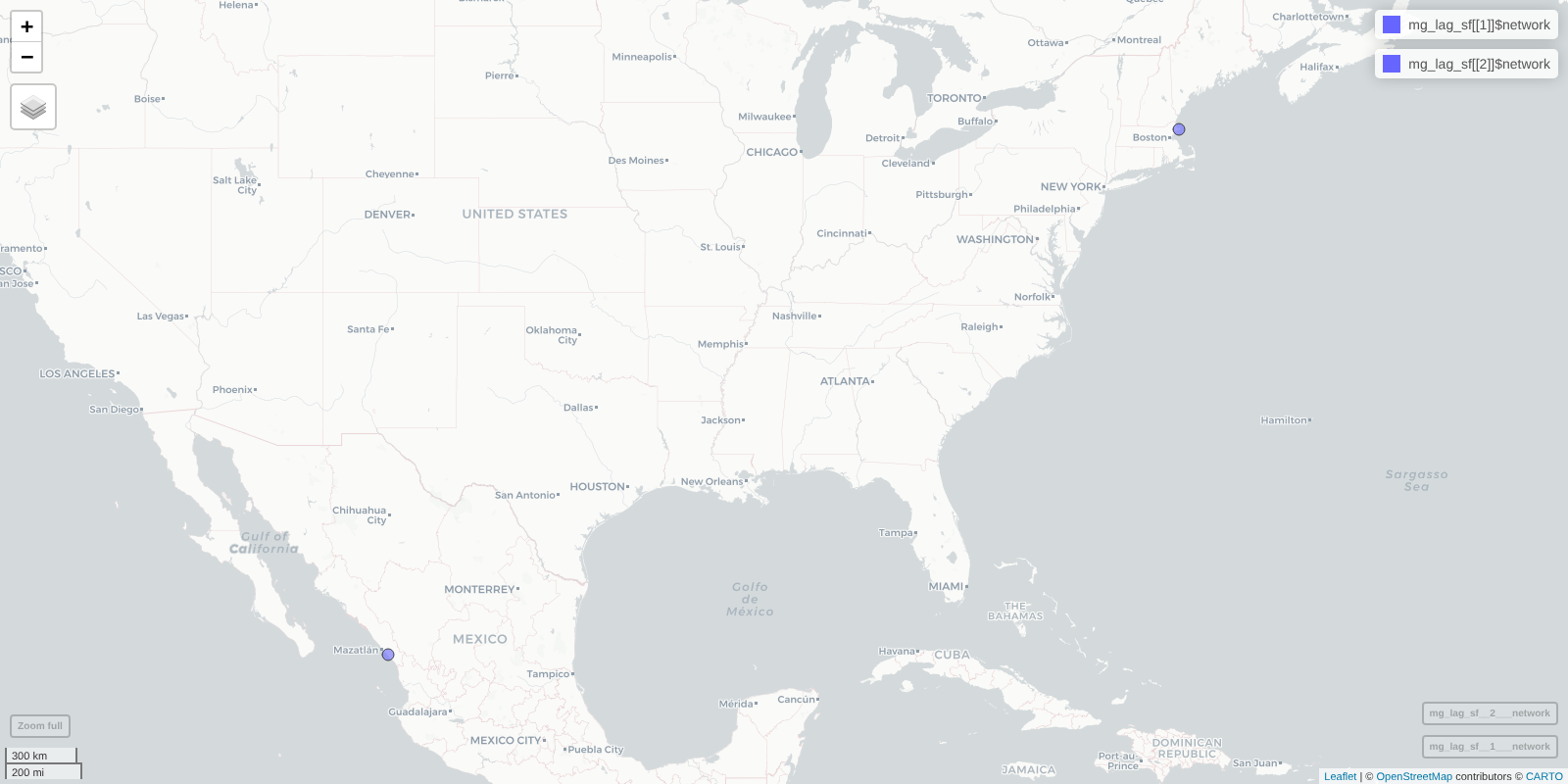
Map showing networks retrieved using the keywork ‘lagoon’
Taxonomic analysis
As Mangal includes taxonomic identifiers, rmangal
can readily be combined with taxize (see taxize for more details
about this package).
Network analysis with igraph
Once the data are retrieved and a mgNetwork or a
mgNetworkCollection objects obtained, it is straightforward
to convert it as a igraph (see the dedicated website) object and then to
carry out network analysis:
library(igraph)
mg_lagoons <- search_datasets(query = 'lagoon') |> get_collection()
# NB the line below returns a list of igraph objects
ig_lagoons <- as.igraph(mg_lagoons)
## Modularity analysis for the first network
modularity(ig_lagoons[[1]], membership(cluster_walktrap(ig_lagoons[[1]])))
#> [1] 0.05139834
## Degree values for all networks
lapply(ig_lagoons, degree)
#> [[1]]
#> 4904 4905 4906 4907 4908 4909 4910 4911 4912 4913 4914 4915 4916 4917 4918 4919 4920 4921 4922 4924 4925 4926 4927 4923 4929 4928
#> 17 11 14 13 18 20 14 10 18 14 12 15 7 15 14 12 14 11 26 7 22 15 21 16 5 17
#>
#> [[2]]
#> 14395 14397 14398 14399 14400 14401 14402 14403 14404 14405 14406 14407 14408 14409 14411 14412 14413 14414 14415 14416 14417 14418 14419
#> 9 5 7 7 5 5 5 4 5 5 5 3 2 4 7 7 5 9 9 5 1 3 5
#> 14420 14422 14423 14424 14425 14426 14427 14396 14428 14410 14421 14429
#> 11 17 4 3 11 11 5 9 5 3 11 2
#>
#> [[3]]
#> 14522 14523 14524 14525 14526 14527 14528 14530 14532 14537 14538 14539 14536 14531 14521 14529 14533 14534 14535
#> 5 5 5 5 5 5 5 9 7 3 2 2 5 9 4 2 7 2 1
#>
#> [[4]]
#> 14430 14431 14432 14433 14434 14435 14436 14437 14438 14439 14440 14441 14442 14443 14444 14445 14446 14448 14449 14450 14451 14452 14453
#> 9 9 5 3 5 8 9 8 9 7 9 3 1 9 8 5 5 12 22 25 23 3 3
#> 14454 14455 14456 14458 14447 14457
#> 3 3 11 8 12 11
#>
#> [[5]]
#> 14496 14497 14498 14499 14500 14501 14502 14503 14504 14505 14506 14507 14508 14511 14512 14513 14515 14516 14517 14518 14519 14520 14509
#> 3 6 6 6 3 3 2 7 7 7 1 1 10 6 8 8 3 3 3 5 5 5 5
#> 14510 14514
#> 6 1
#>
#> [[6]]
#> 14459 14460 14461 14462 14463 14464 14465 14466 14467 14468 14469 14471 14472 14473 14474 14475 14476 14477 14478 14479 14480 14481 14482
#> 12 12 1 6 6 15 15 15 15 6 4 9 9 9 13 13 13 2 2 14 14 12 17
#> 14483 14485 14486 14487 14488 14491 14493 14494 14495 14492 14490 14470 14484 14489
#> 17 17 12 12 7 7 2 2 2 7 7 4 17 3Network manipulation and visualization with tidygraph
and ggraph
The package tidygraph
treats networks as two tidy tables (one for the edges and one for the
nodes) that can be modified using the grammar of data manipulation
developed in the tidyverse.
Moreover, tidygraph wraps over most of the
igraph functions so that the user can call a vast variety
of algorithms to properly analysis networks. Fortunately, objects of
class mgNetwork can readily be converted into
tbl_graph objects which allows the user to benefit from all
the tools included in tidygraph:
library(tidygraph)
# NB the line below would not work with a mgNetworksCollection (use lapply)
tg_lagoons <- as_tbl_graph(mg_lagoons[[1]]) |>
mutate(centrality_dg = centrality_degree(mode = 'in'))
tg_lagoons %E>% as_tibble()
#> # A tibble: 189 × 19
#> from to interaction_id date direction type method attr_id value public network_id created_at updated_at attribute.id attribute.name
#> <int> <int> <int> <chr> <chr> <chr> <lgl> <int> <dbl> <lgl> <int> <chr> <chr> <int> <chr>
#> 1 9 9 48376 2003-01… directed pred… NA 12 0.026 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 2 9 11 48377 2003-01… directed pred… NA 12 0.025 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 3 9 12 48378 2003-01… directed pred… NA 12 0.003 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 4 9 15 48379 2003-01… directed pred… NA 12 0.009 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 5 9 16 48380 2003-01… directed pred… NA 12 0.009 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 6 9 17 48381 2003-01… directed pred… NA 12 0.016 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 7 9 18 48382 2003-01… directed pred… NA 12 0.284 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 8 9 19 48383 2003-01… directed pred… NA 12 0.231 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 9 9 21 48384 2003-01… directed pred… NA 12 0.079 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> 10 9 22 48385 2003-01… directed pred… NA 12 0.09 TRUE 86 2019-02-2… 2019-02-2… 12 dietary matrix
#> # ℹ 179 more rows
#> # ℹ 4 more variables: attribute.description <chr>, attribute.unit <lgl>, attribute.created_at <chr>, attribute.updated_at <chr>
tg_lagoons %N>% as_tibble() |>
select(original_name, taxonomy.tsn, centrality_dg)
#> # A tibble: 26 × 3
#> original_name taxonomy.tsn centrality_dg
#> <chr> <int> <dbl>
#> 1 Scianids 169237 1
#> 2 Elopids 28630 0
#> 3 Lutjanids 168845 1
#> 4 Carangids 168584 2
#> 5 Centropomids 167642 2
#> 6 Ariids 43998 1
#> 7 Haemulids 169055 4
#> 8 Pleuronectoids 172859 3
#> 9 Callinectes 13951 6
#> 10 Belonoids 165546 4
#> # ℹ 16 more rowsAnother strong advantage of tbl_graph objects is that
there are the objects used by the package ggraph that
that offers various functions (theme, geoms, etc.) to efficiently
visualize networks:
library(ggraph)
ggraph(tg_lagoons, layout = "stress") +
geom_edge_parallel(
end_cap = circle(.5),
start_cap = circle(.5),
arrow = arrow(length = unit(1, 'mm'),
type = 'closed')
) +
geom_node_point(aes(colour = taxonomy.rank), size = 8) +
theme_graph(background = "grey40", foreground = NA, text_colour = 'white')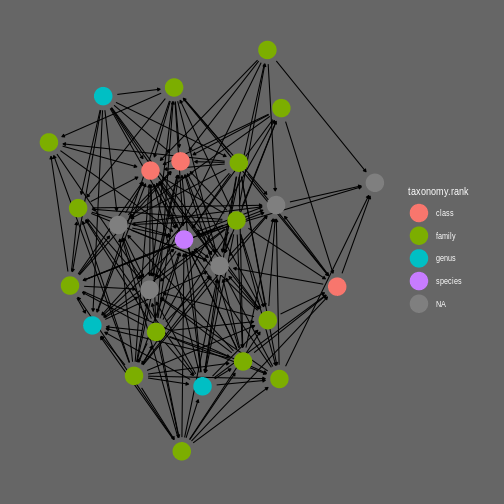
Example of network visualization with ggraph
Creating a list references for a set of networks
We can easily print the BibTeX of all publications involved in the networks collection.
search_datasets(query = 'lagoon') |>
get_collection() |>
get_citation() |>
cat(sep = "\n\n")
#> @article{Zetina_Rej_n_2003, doi = {10.1016/s0272-7714(02)00410-9}, url = {https://doi.org/10.1016%2Fs0272-7714%2802%2900410-9}, year = 2003, month = {aug}, publisher = {Elsevier {BV}}, volume = {57}, number = {5-6}, pages = {803--815}, author = {Manuel J. Zetina-Rejón and Francisco Arreguí-Sánchez and Ernesto A. Chávez}, title = {Trophic structure and flows of energy in the Huizache{ extendash}Caimanero lagoon complex on the Pacific coast of Mexico},journal = {Estuarine, Coastal and Shelf Science}}
#>
#> @article{Dexter_1947, doi = {10.2307/1948658}, url = {https://doi.org/10.2307%2F1948658}, year = 1947, month = {feb}, publisher = {Wiley}, volume = {17}, number = {3}, pages = {261--294}, author = {Ralph W. Dexter}, title = {The Marine Communities of a Tidal Inlet at Cape Ann, Massachusetts: A Study in Bio-Ecology}, journal = {Ecological Monographs}}