refsplitr: author name disambiguation, author georeferencing, and mapping of coauthorship networks with Web of Science data.
refsplitr is an R package to parse and organize reference records downloaded from the Web of Science citation database, disambiguate the names of authors, geocode their locations, and generate/visualize coauthorship networks. The Web of Science (WOS) is a toll-access literature and citation database maintained by Clarivate Analytics that indexes articles from ~12,000 academic journals. WOS records include a diversity of data about each article (e.g., article title, journal name, author names, author addresses, number of times the article has been cited, funding sources), making them very useful for studying patterns of scientific productivity, coauthorship, research impact, and other Science of Science topics. Because bulk WOS records and API access to the WOS are very expensive, researchers at WOS-subscribing institutions often gather data by conducting WOS searches and downloading reference records in batches. However, this requires a cumbersome process of extracting, merging, and correcting data from the downloaded records prior to conducting any analyses. refsplitr will rapidly merge and process reference data files downloaded from the WOS, and then process and organize them in a format amenable for use in scientometric, social network, and Science of Science analyses.
Support for the development of refsplitr was provided by grants from the University of Florida Center for Latin American Studies and the University of Florida Informatics Institute.
Installation
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("ropensci/refsplitr")Workflow
There are four steps in the refsplitr package’s workflow:
- Importing and tidying Web of Science reference records (be sure to download records using the procedure in Appendix 1 of the vignette)
- Author name disambiguation and parsing of author addresses
- Georeferencing of author institutions using either the Nominatim service, which uses OpenStreetMap data and which
refsplitrqueries via the [tidygeocoder]((https://jessecambon.github.io/tidygeocoder/) package (default; free) OR the Data Science Toolkit, which uses the Google maps API (limited number of free queries after which users must pay); for additional details on pricing information how to register with Google to use their API see therefsplitrvignette. - Data visualization
The procedures required for these four steps,each of which is implemented with a simple command, are described in detail in the refsplitr vignette. An example of this workflow is provided below:
load the Web of Science records into a dataframe
dat1 <- references_read(data = system.file("extdata", "example_data.txt", package = "refsplitr"), dir = FALSE) disambiguate author names and parse author address
dat2 <- authors_clean(references = dat1)after reviewing disambiguation, merge any necessary corrections
dat3 <- authors_refine(dat2$review, dat2$prelim)georeference the author locations
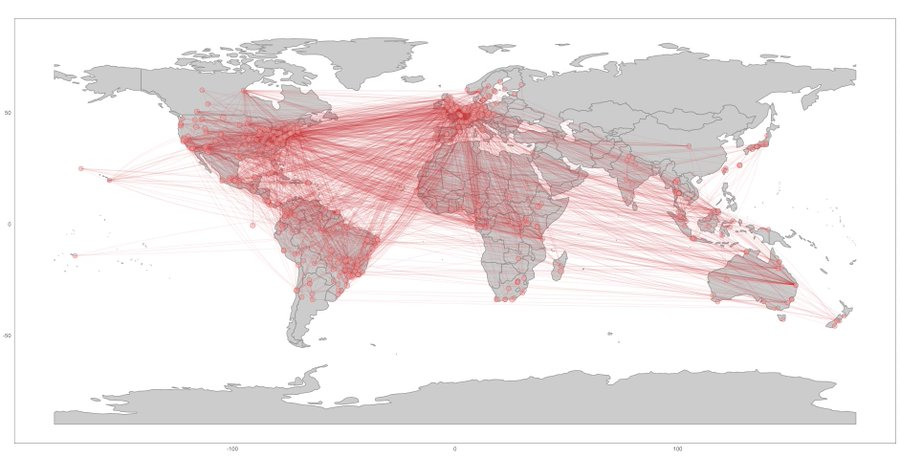
dat4 <- authors_georef(dat3)generate a map of coauthorships; this is only one of the five possible visualizations
plot_net_address(dat4$addresses) Registering with Google for an API key (if using Google Maps to georeference addresses)
- Install and load the
ggmappackage
install.packages("ggmap")
library(ggmap)Register for a Google Geocoding API by following the instructions on the
READ MEof theggmaprepository.Once you have your API key, add it to your
~/.Renvironwith the following:
ggmap::register_google(key = "[your key]", write = TRUE)- You should now be able to use
authors_georef()as described in the vignette. WARNING:refsplitrcurrently has a limit of 2500 API calls per day. We are working on including the ability for users to select their own limits.
Remember: Your API key is unique and for you alone. Don’t share it with other users or record it in a script file that is saved in a public repository. If need be you can visit the same website where you initially registered and generate a new key.
Improvements & Suggestions
We welcome any suggestions for package improvement or ideas for features to include in future versions. If you have Issues, Feature Requests and Pull Requests, here is how to contribute. We expect everyone contributing to the package to abide by our Code of Conduct.
Contributors
- Auriel Fournier, Porzana Solutions
- Matt Boone, Porzana Solutions
- Forrest Stevens, University of Louisville
- Emilio M. Bruna, University of Florida
Citation
The refsplitr package has been described in an article in the Journal of Open Source Software. We request that you cite both the package and the publication when using refsplitr in your work.
Citation: refsplitr Package
Auriel M.V. Fournier, Matthew E. Boone, Forrest R. Stevens, and Emilio M. Bruna Developer (2025). refsplitr: author name disambiguation, author georeferencing, and mapping of coauthorship networks with Web of Science data. R package version 1.2.0. https://github.com/ropensci/refsplitr
@Manual{refsplitr2025,
title = {refsplitr: author name disambiguation, author georeferencing,
and mapping of coauthorship networks with Web of Science data.},
author = {Fournier, Auriel M.V., Matthew E. Boone, Forrest R. Stevens, and Emilio M. Bruna},
year = {2020},
note = {R package version 1.2.0.},
url={https://github.com/ropensci/refsplitr}
}Citation: JOSS paper
Fournier et al., (2020). refsplitr: Author name disambiguation, author georeferencing, and mapping of coauthorship networks with Web of Science data. Journal of Open Source Software, 5(45), 2028, https://doi.org/10.21105/joss.02028
@article{Fournier2020,
doi = {10.21105/joss.02028},
url = {https://doi.org/10.21105/joss.02028},
year = {2020}, publisher = {The Open Journal},
volume = {5},
number = {45},
pages = {2028},
author = {Auriel M.v. Fournier and Matthew E. Boone and Forrest R. Stevens and Emilio M. Bruna},
title = {refsplitr: Author name disambiguation, author georeferencing, and mapping of coauthorship networks with Web of Science data},
journal = {Journal of Open Source Software}
}License
Note regarding package development history: The early development of refsplitr- initially known as refnet - was by Forrest Stevens and Emilio M. Bruna and was done on r-forge. In December 2017 Bruna moved it to Github and hired Porzana Solutions to finalize the package and prepare it for submission to rOpenSci. Please make all suggestions for changes via this Github repository - do not make a repo mirror of the R-forge version.
